GPSAdb 2.0 — Atlas of Gene-Perturbation Transcriptomes
GPSAdb 2.0 is a comprehensive, open resource for exploring transcriptomic consequences of gene perturbations in human cell lines. The database hosts 7,665 perturbation groups (42,235 RNA-seq samples) covering 2,810 targeted genes across 1,063 cell lines, uniformly processed and richly annotated for cross-study analysis.
What you can do with GPSAdb 2.0
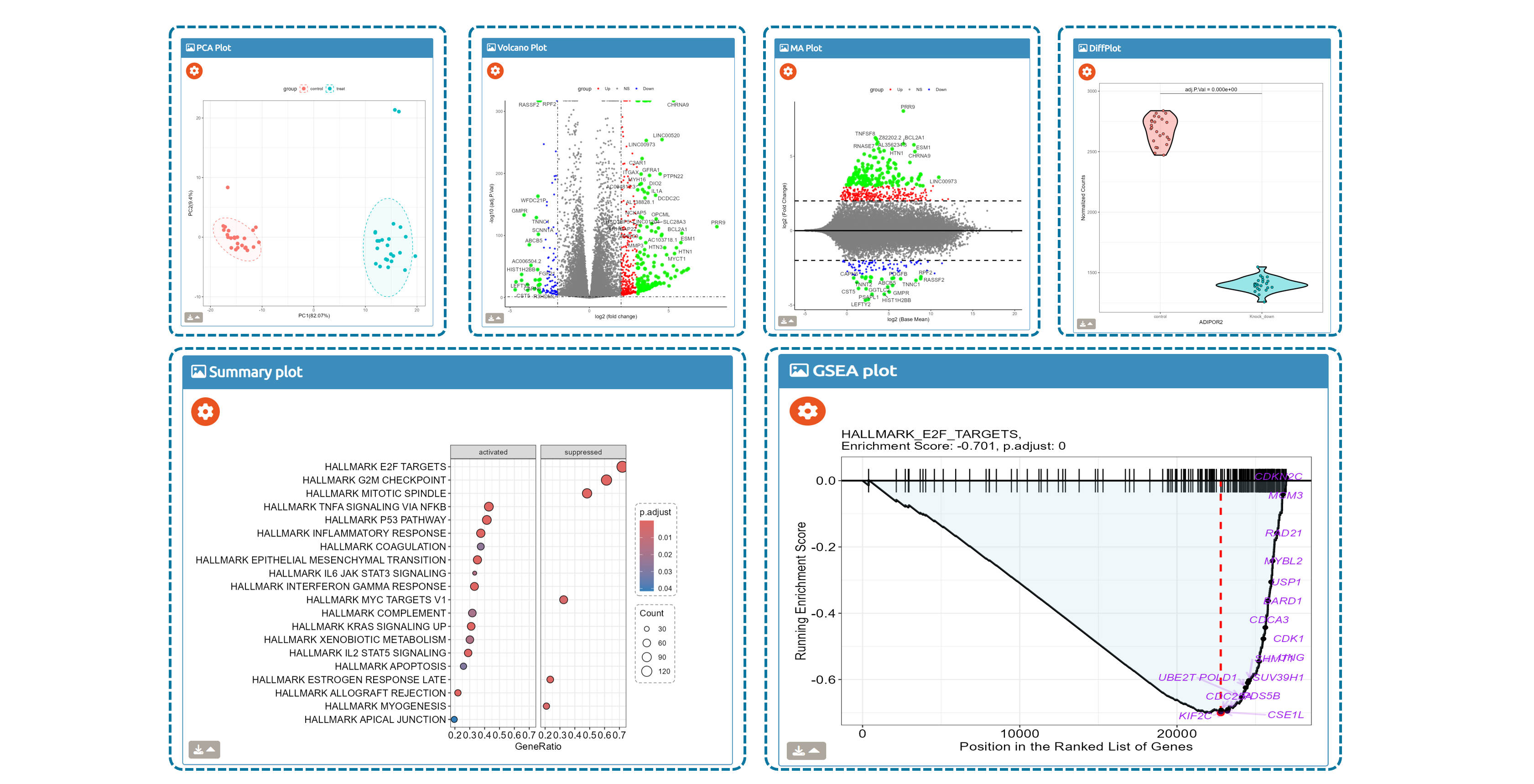
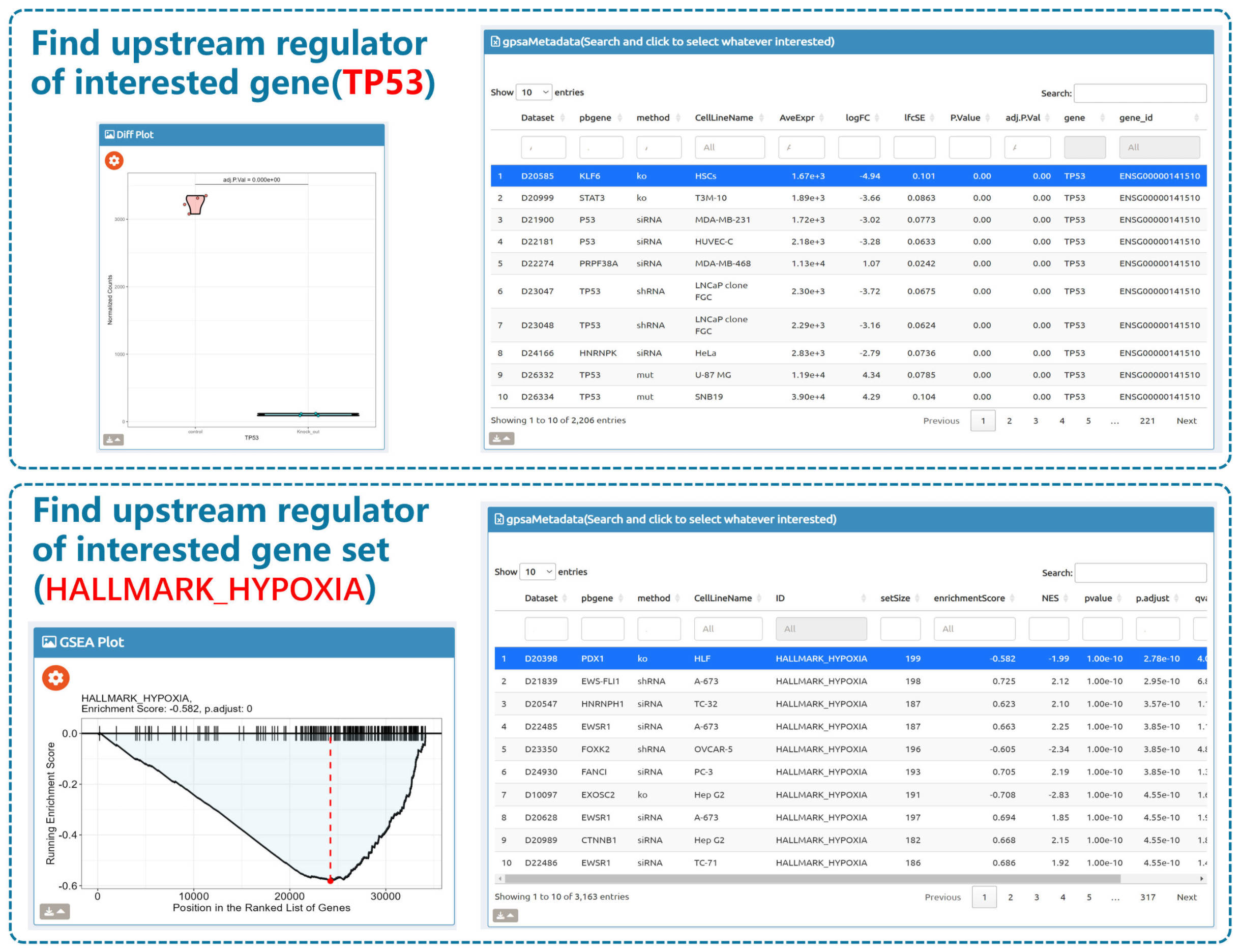
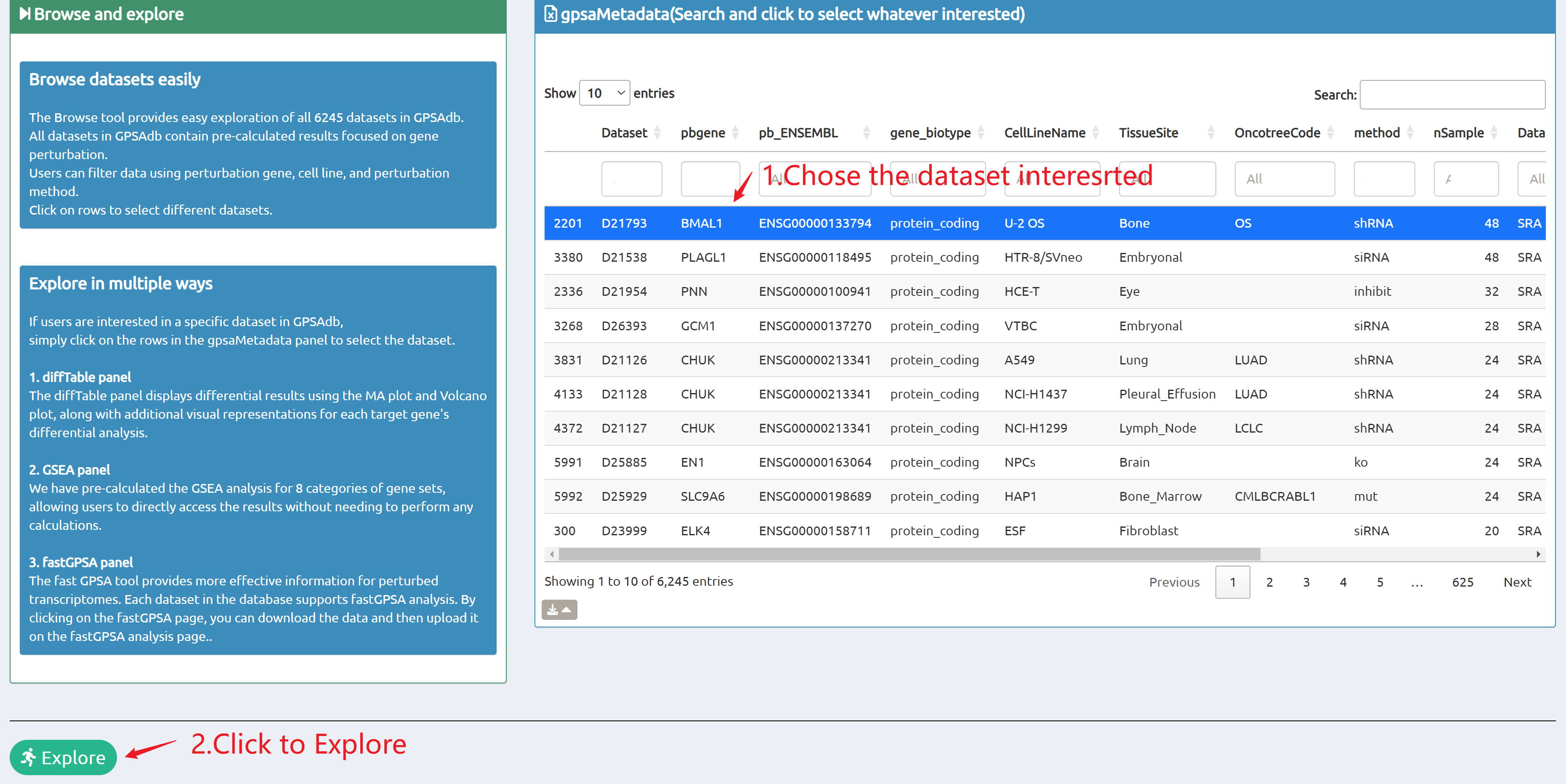
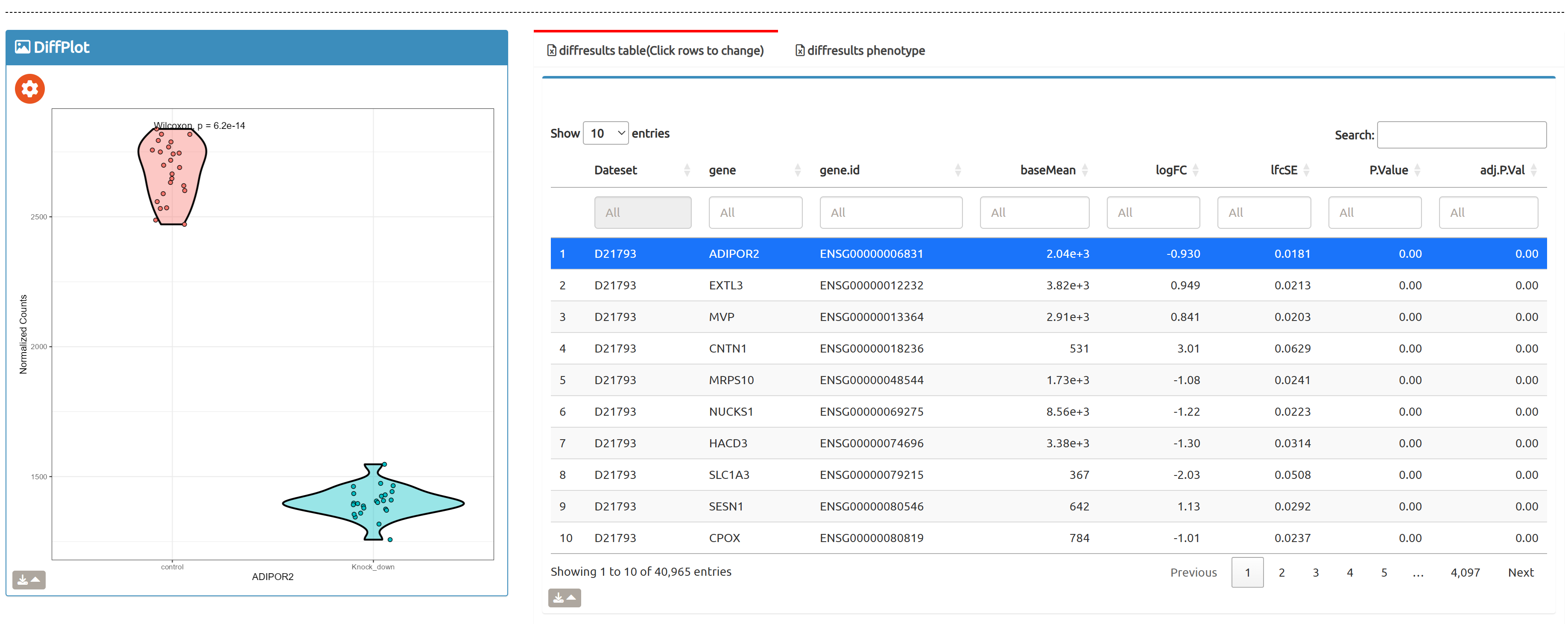
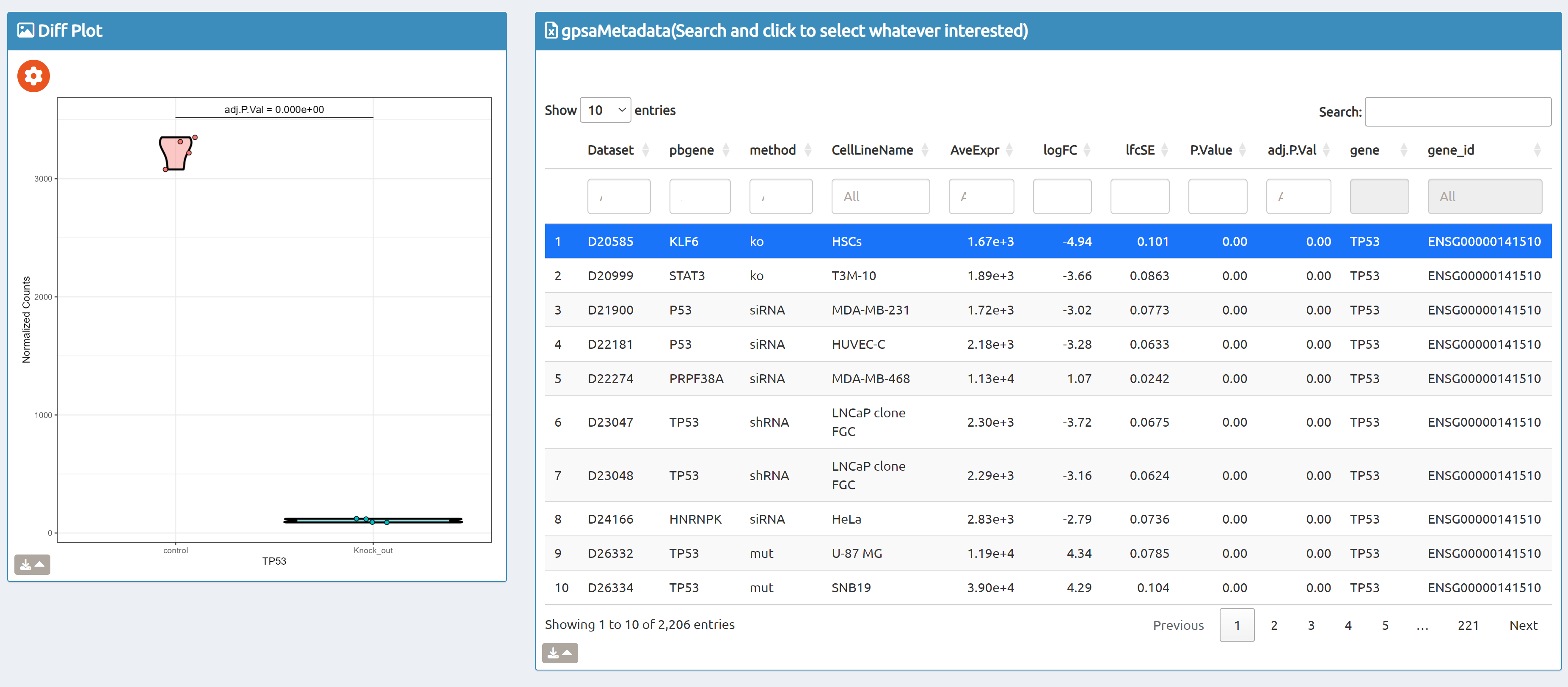
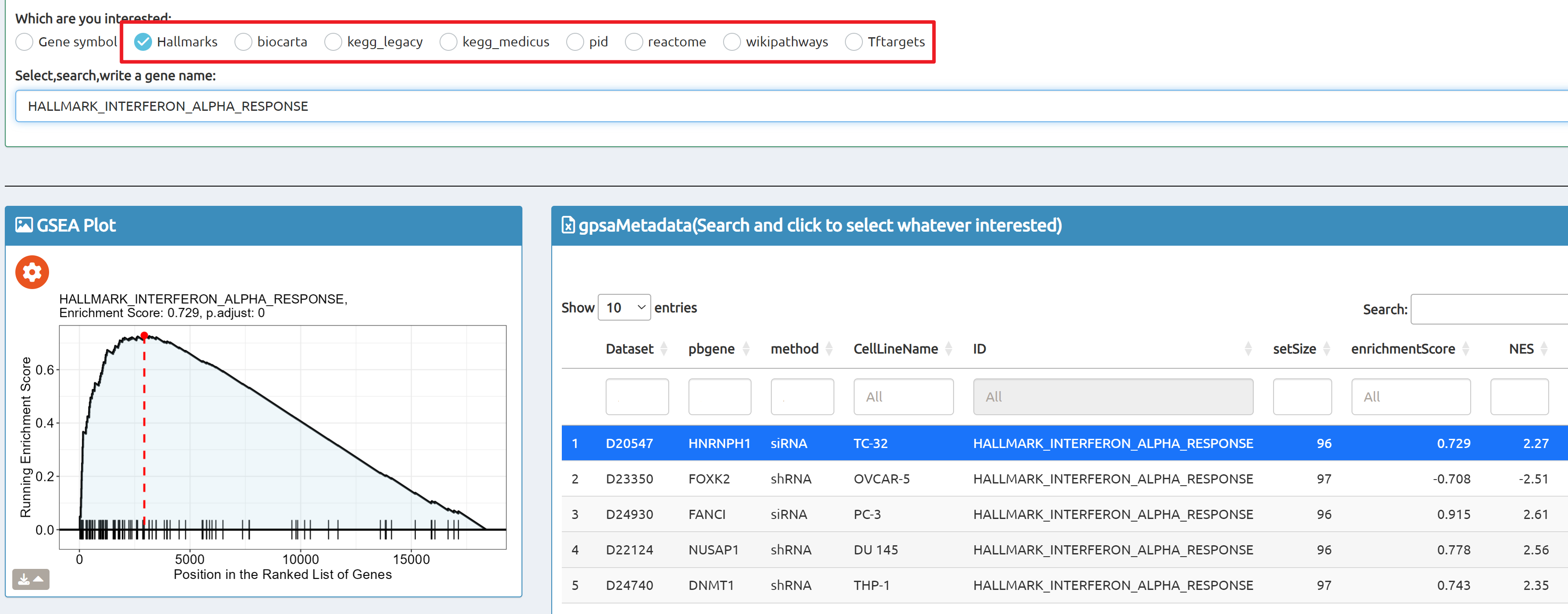
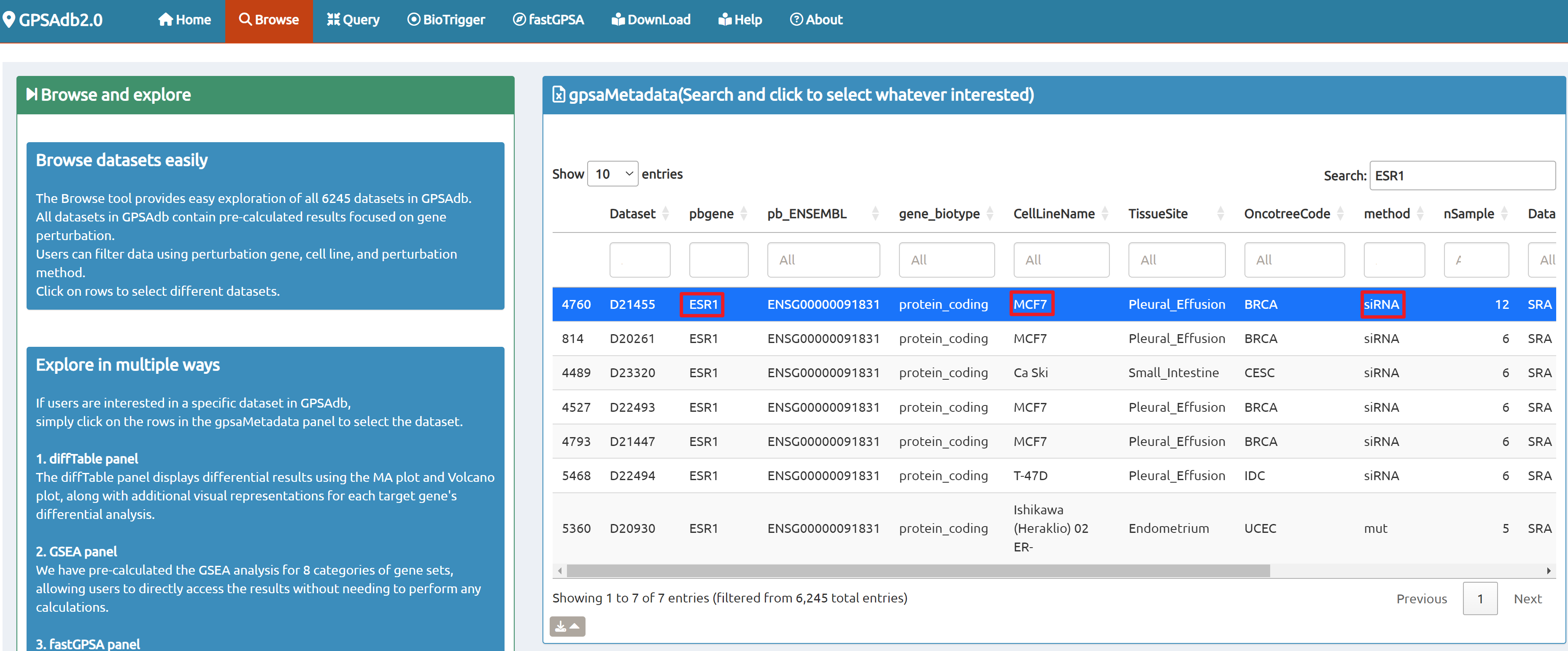
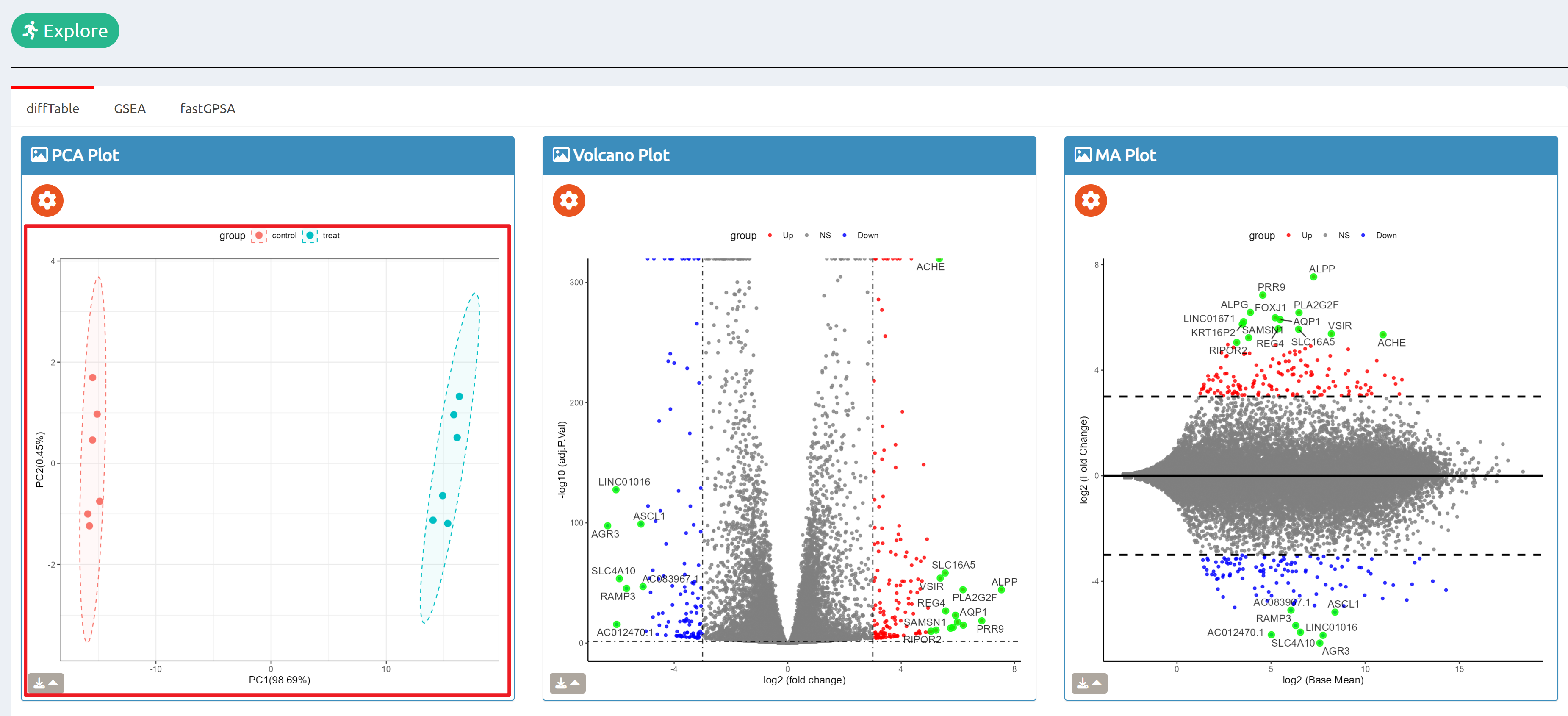
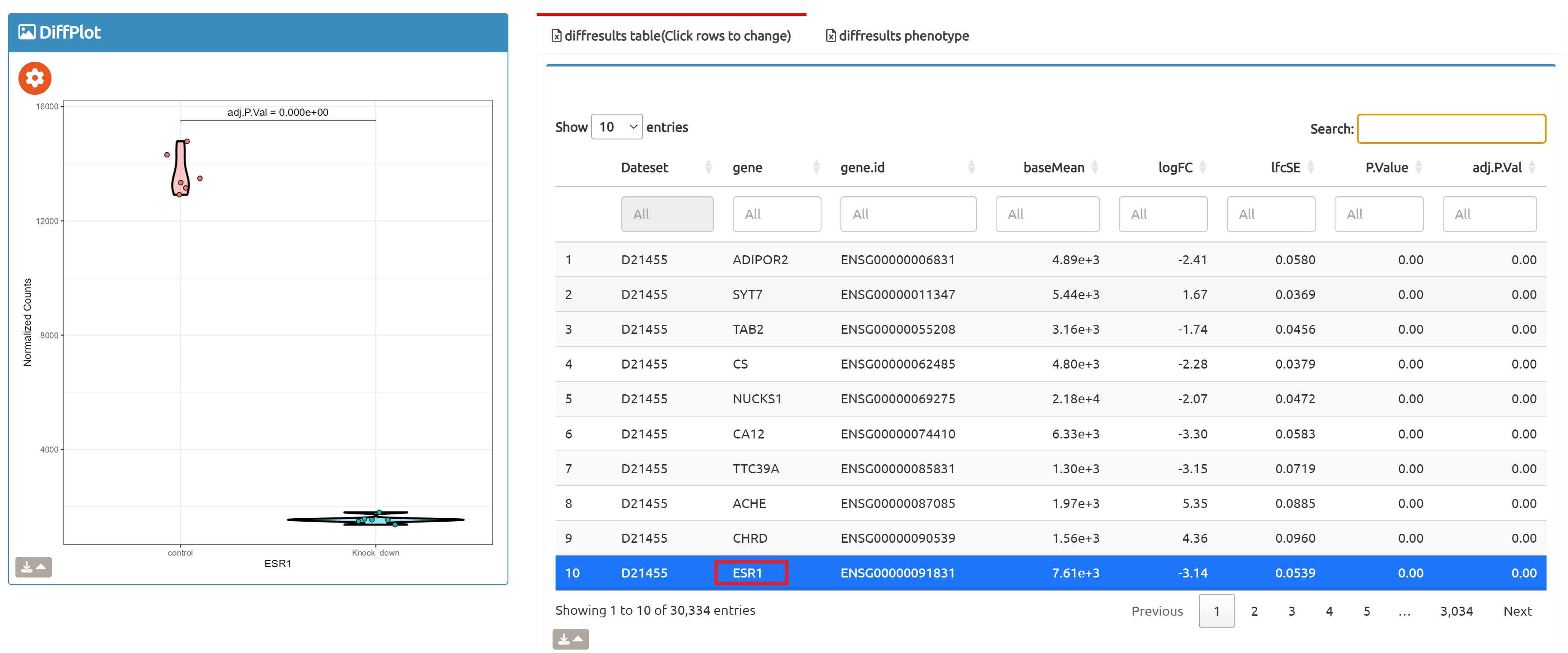
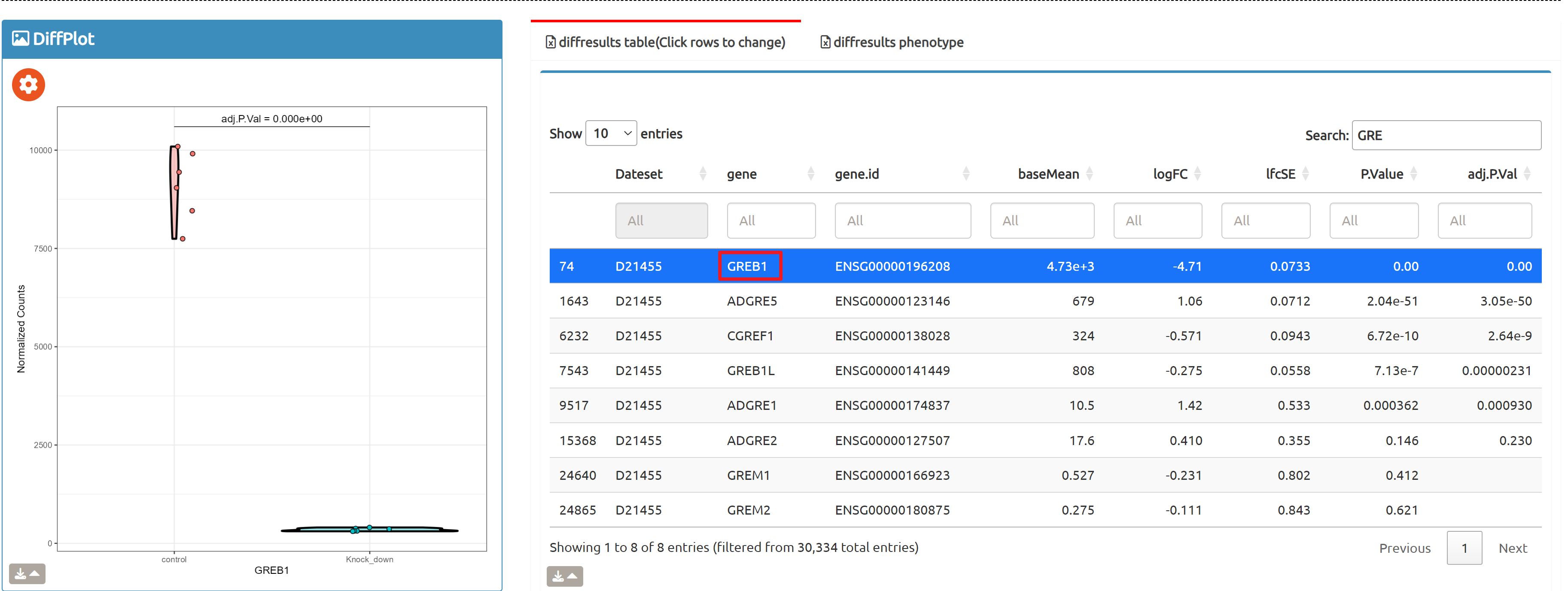
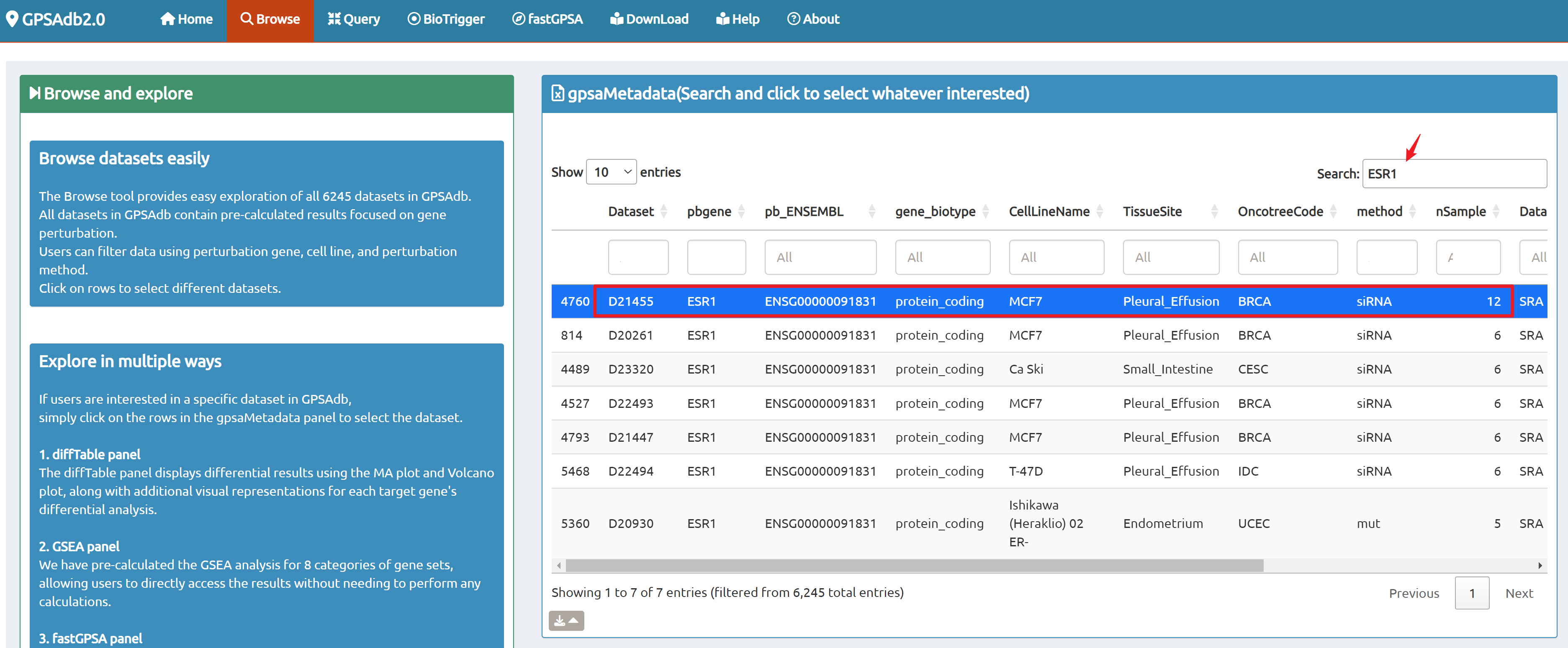
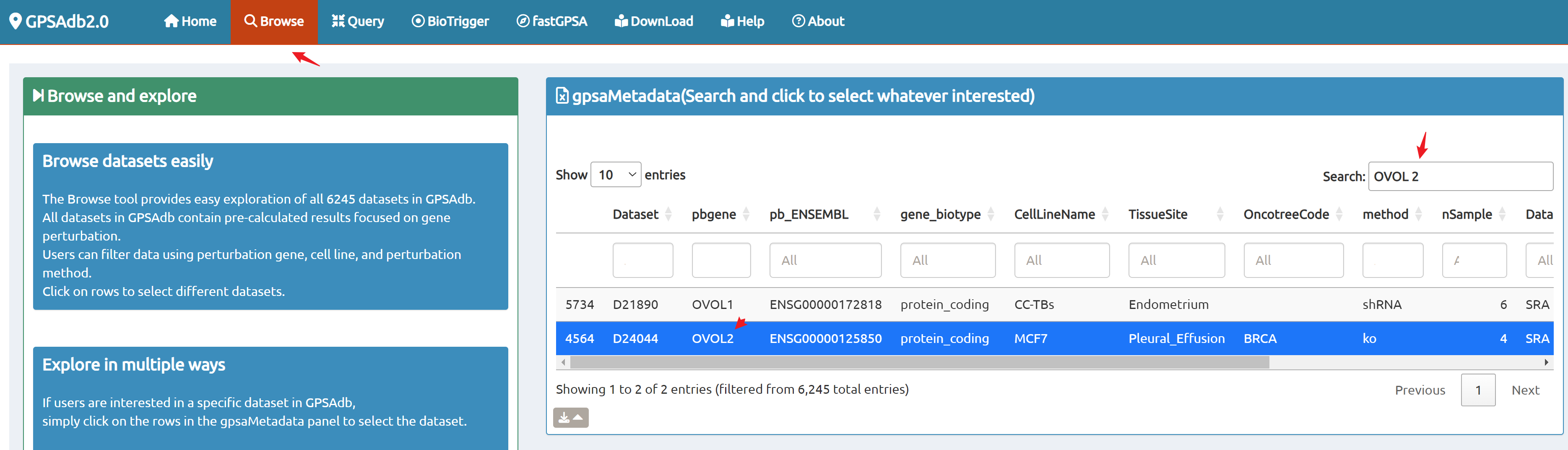
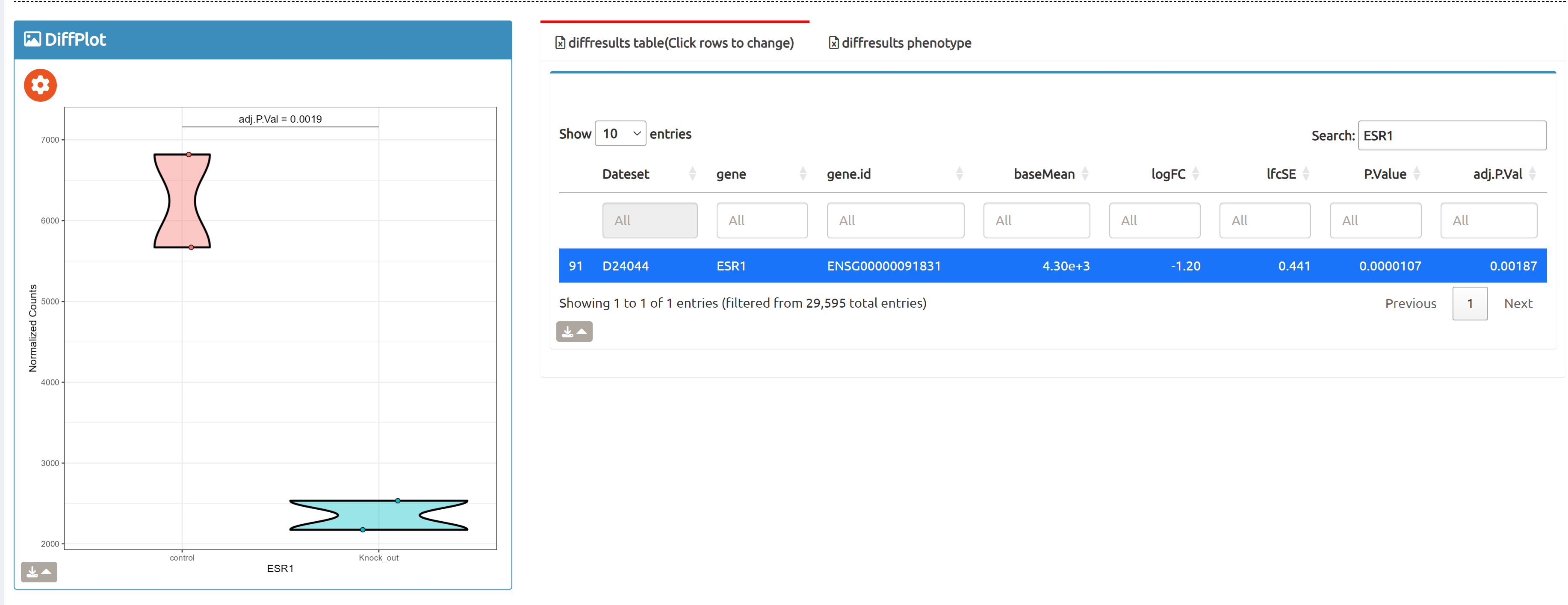
- Browse perturbation biology: Explore 15,330 perturbation gene sets and drill down by gene, cell line, or pathway.
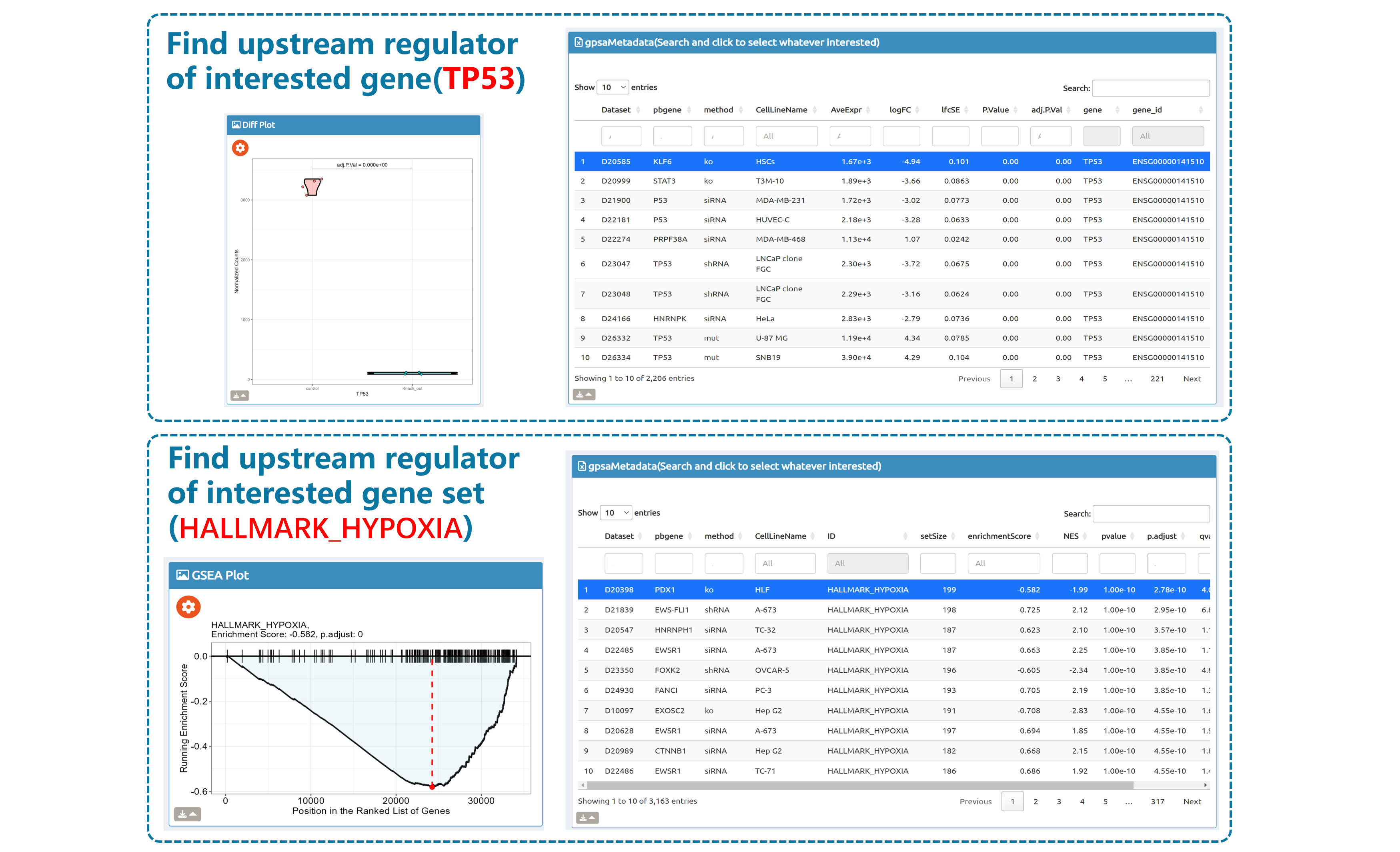
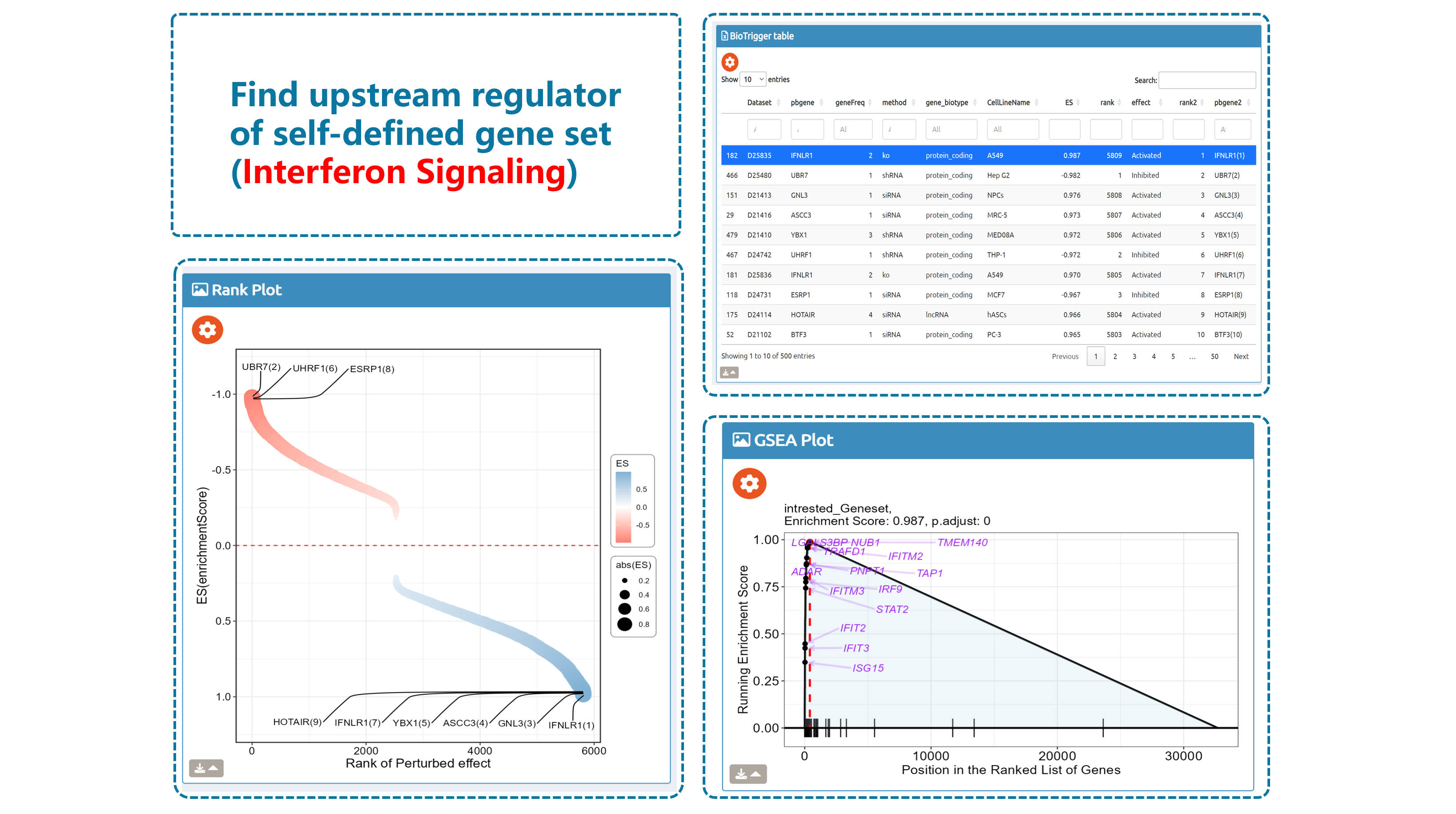
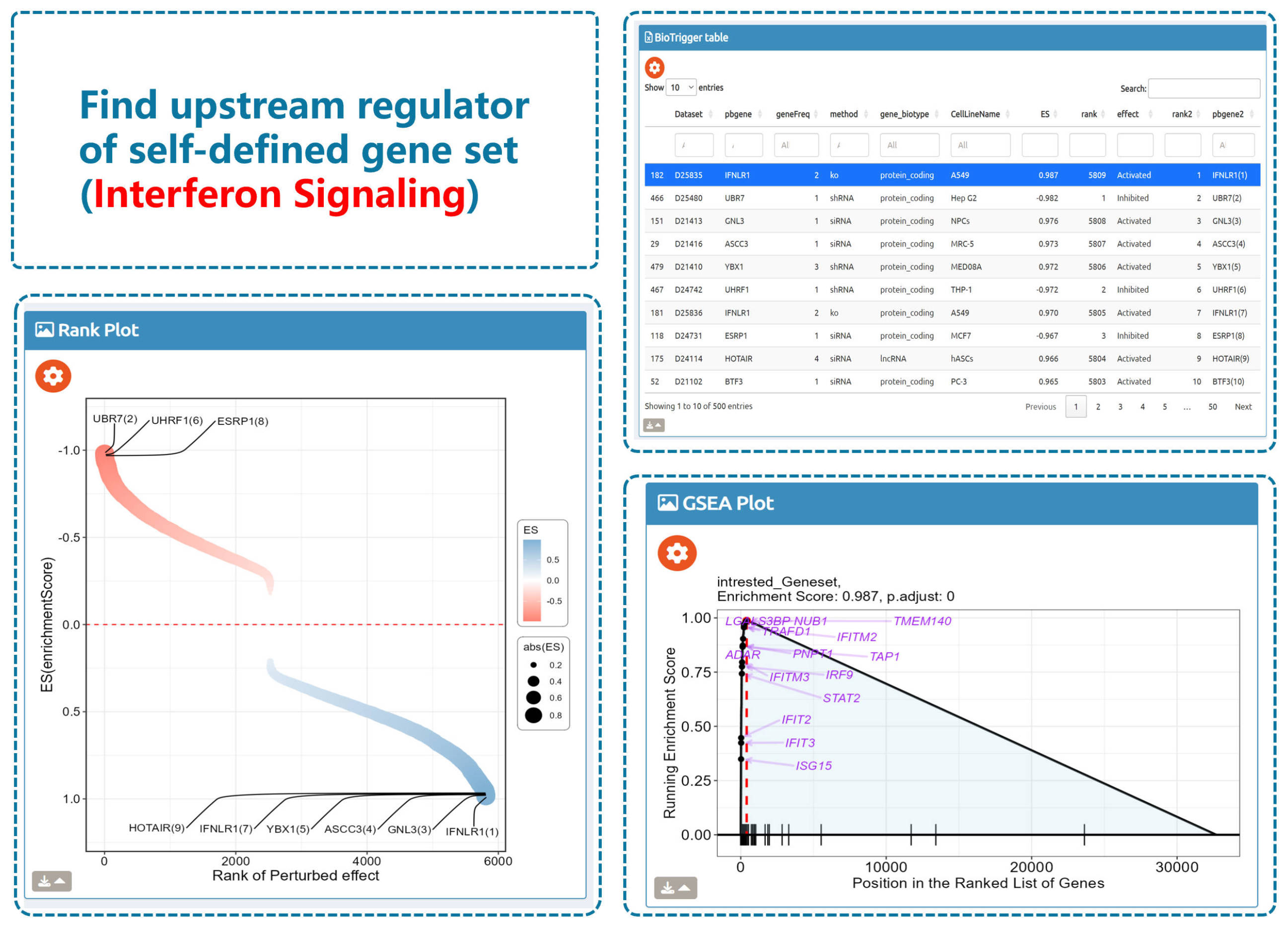
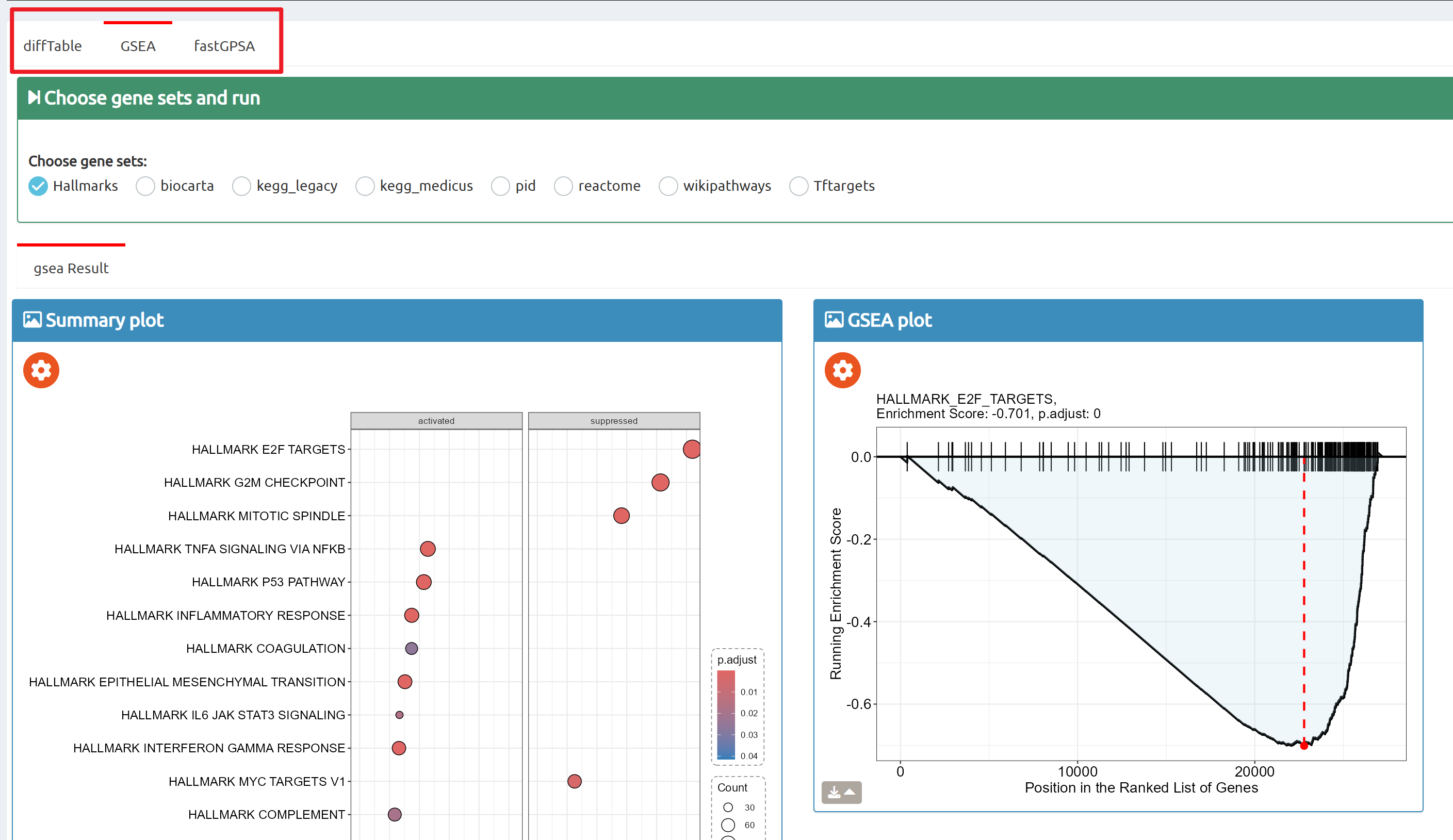
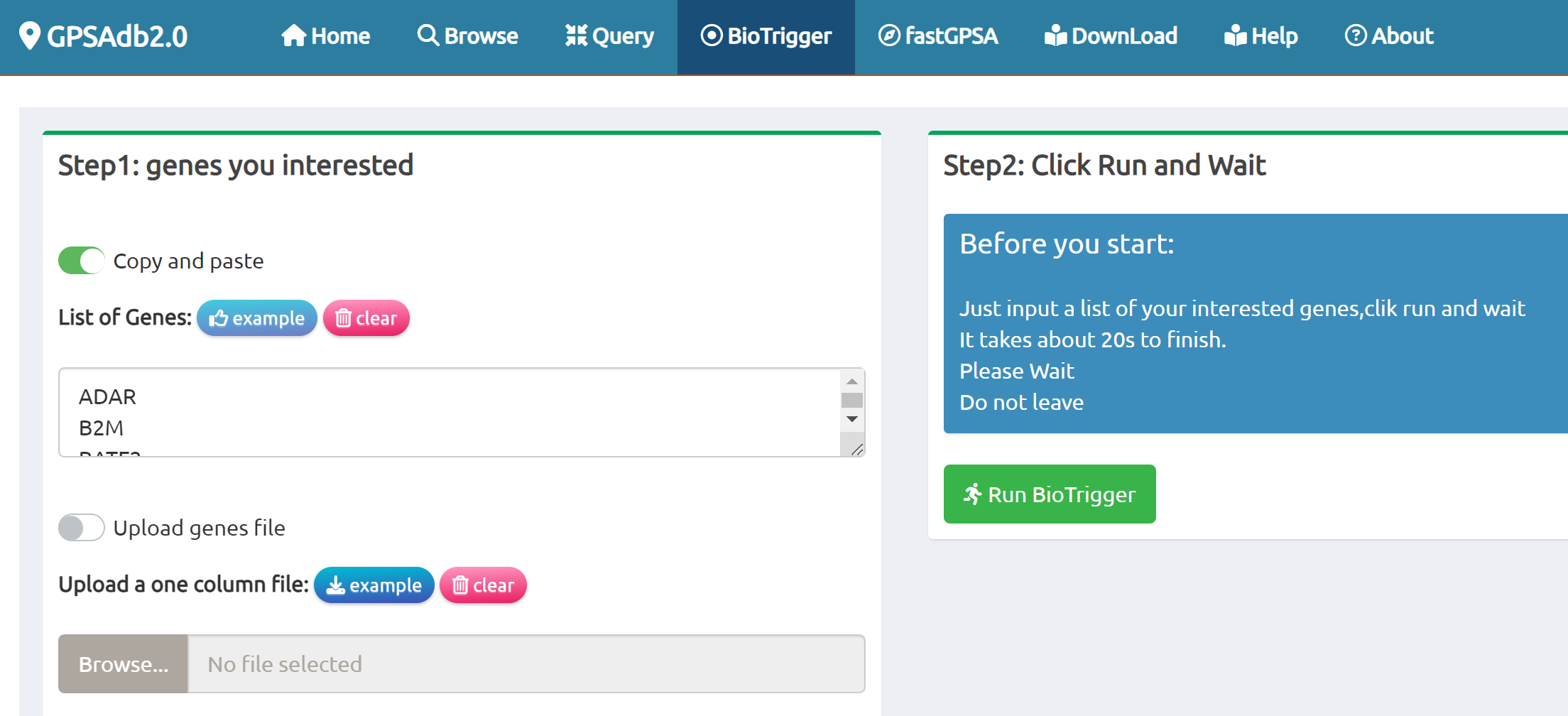
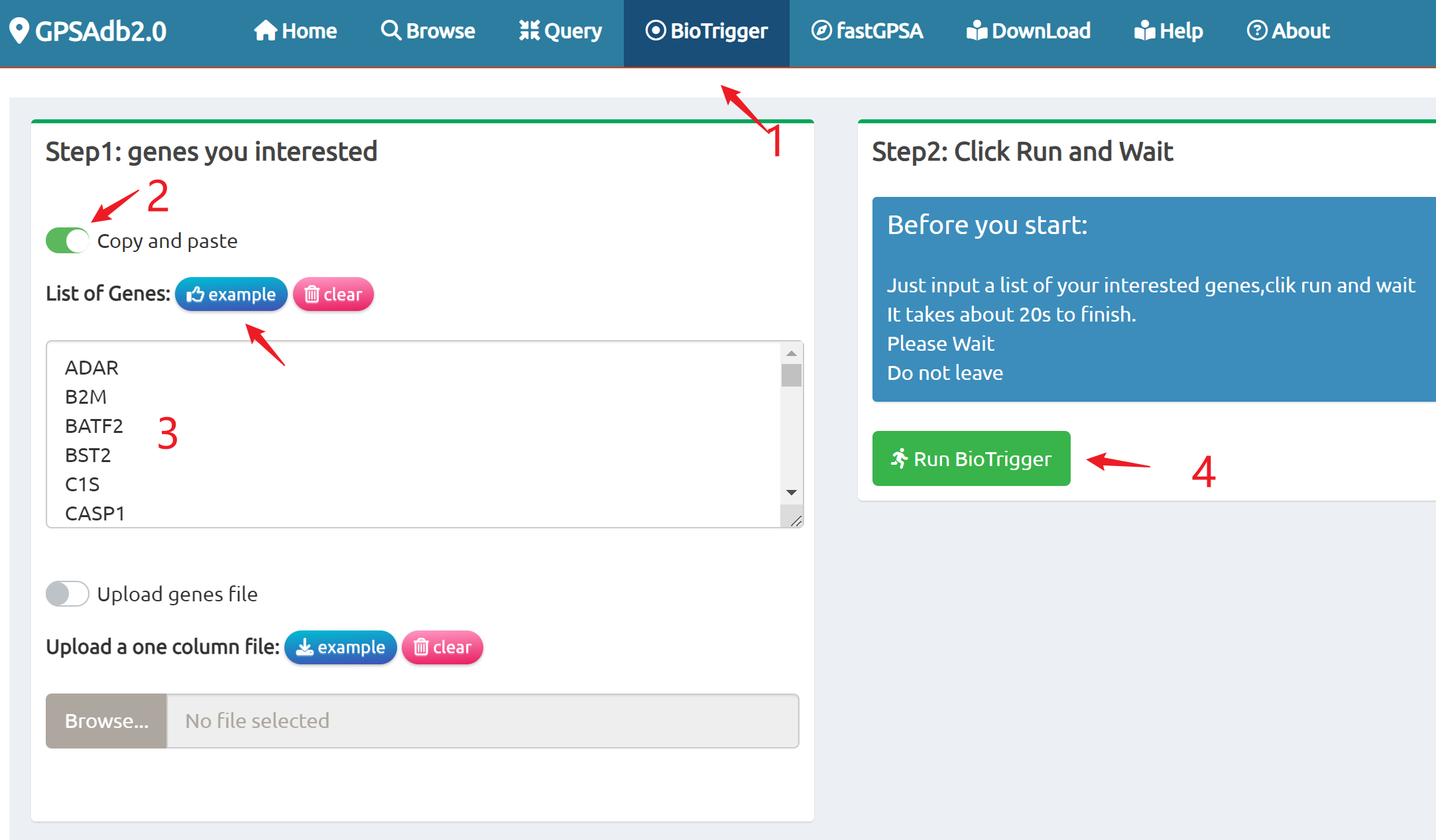
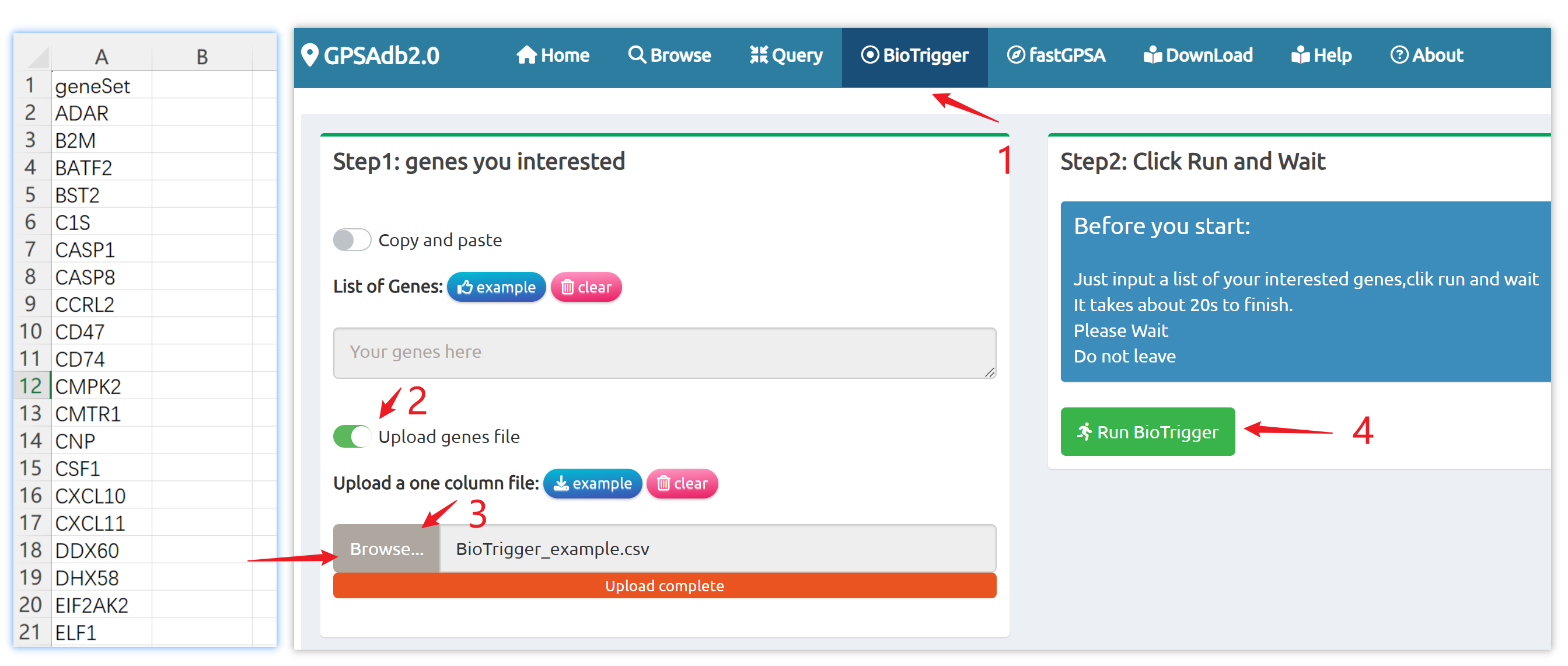
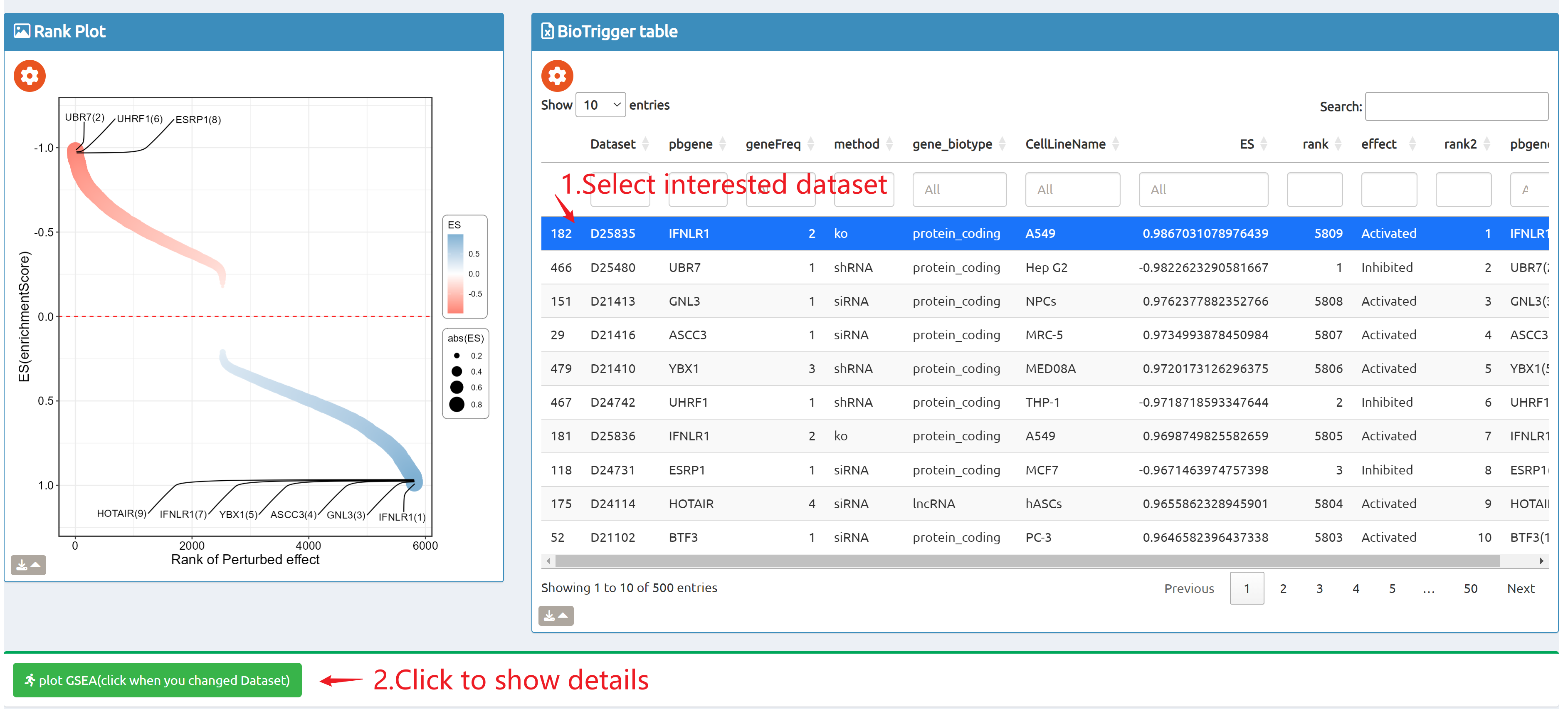
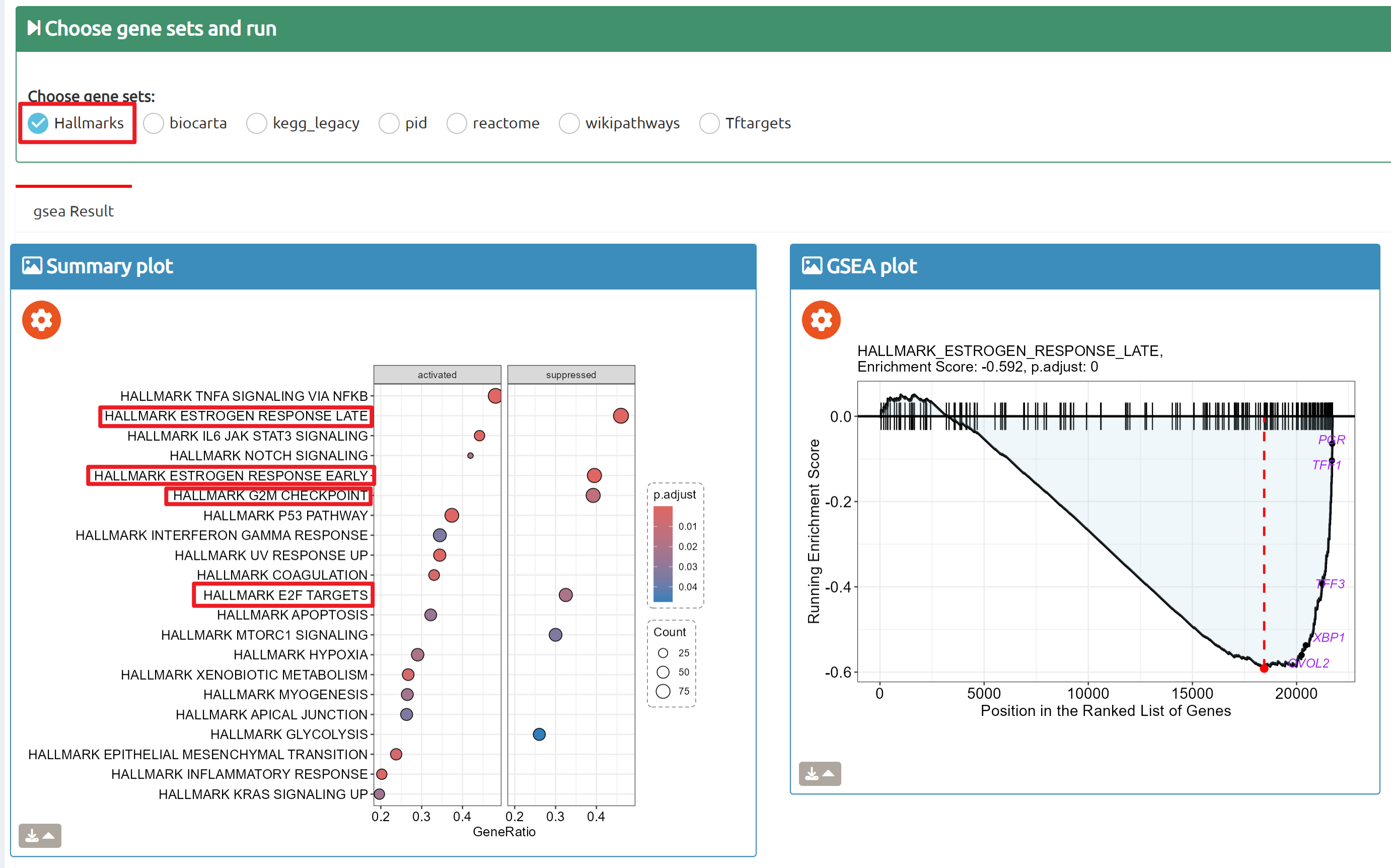
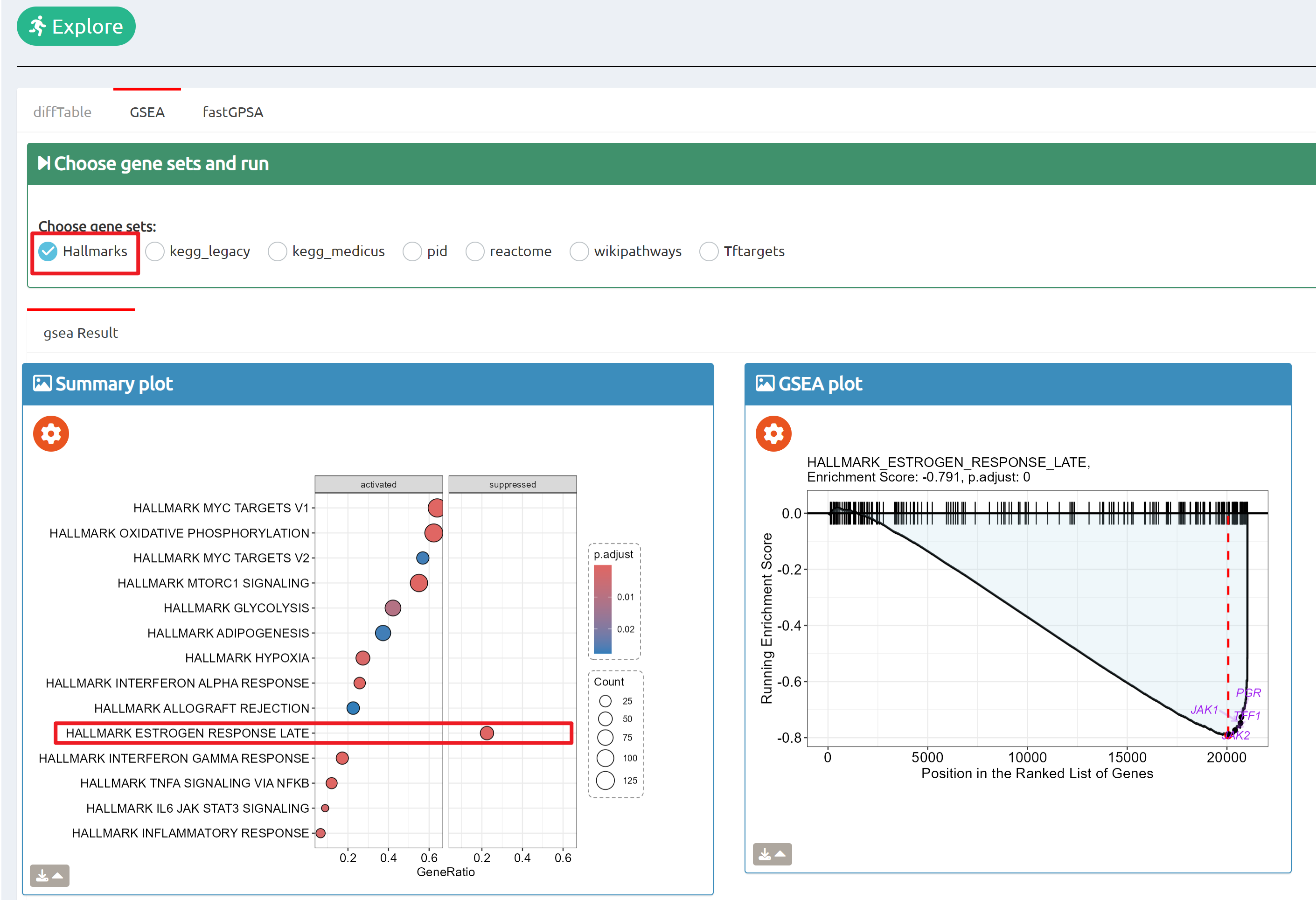
- BioTrigger: Run flexible GSEA on 5,809 curated datasets using a single or paired gene set to identify upstream regulators of your biology of interest.
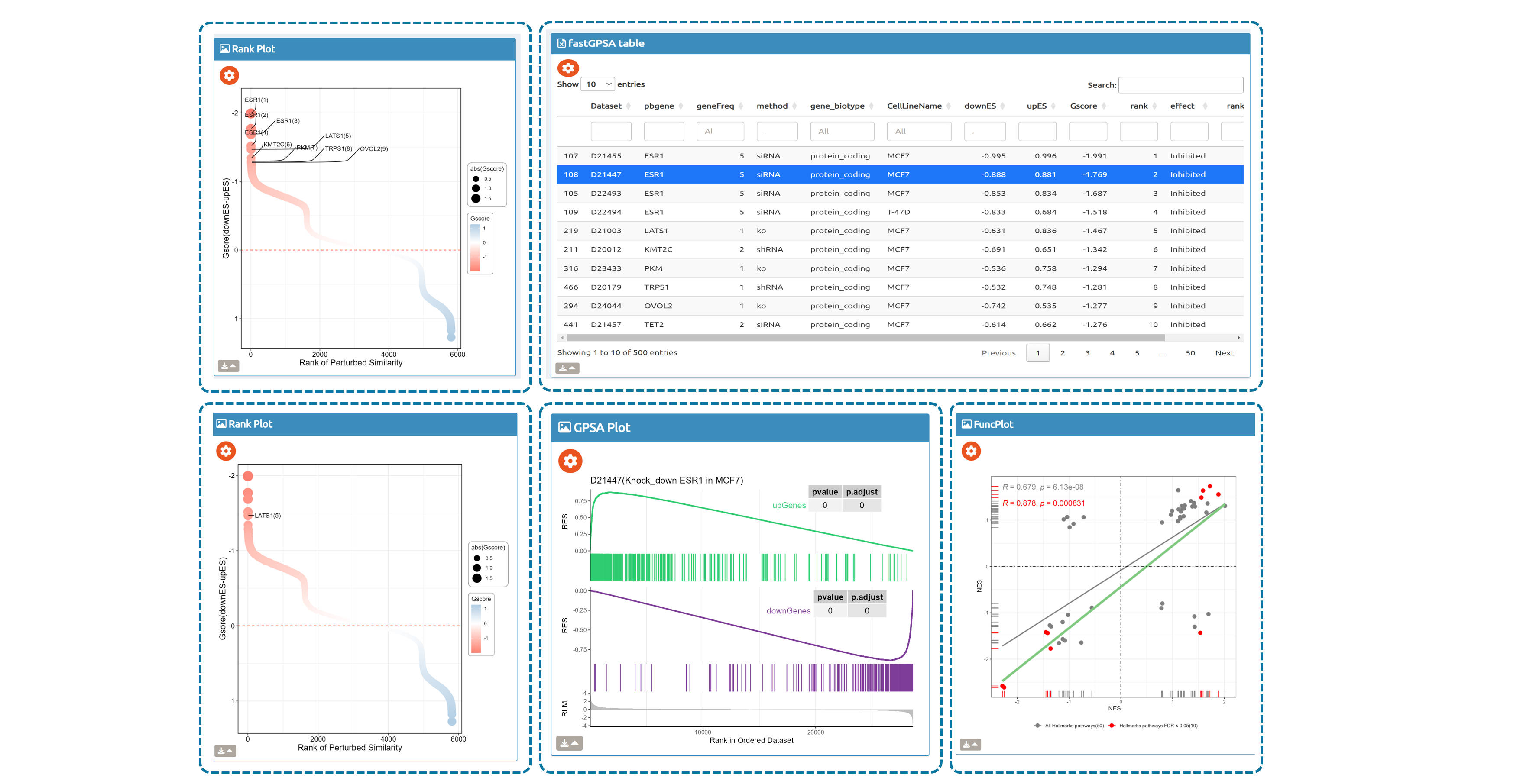
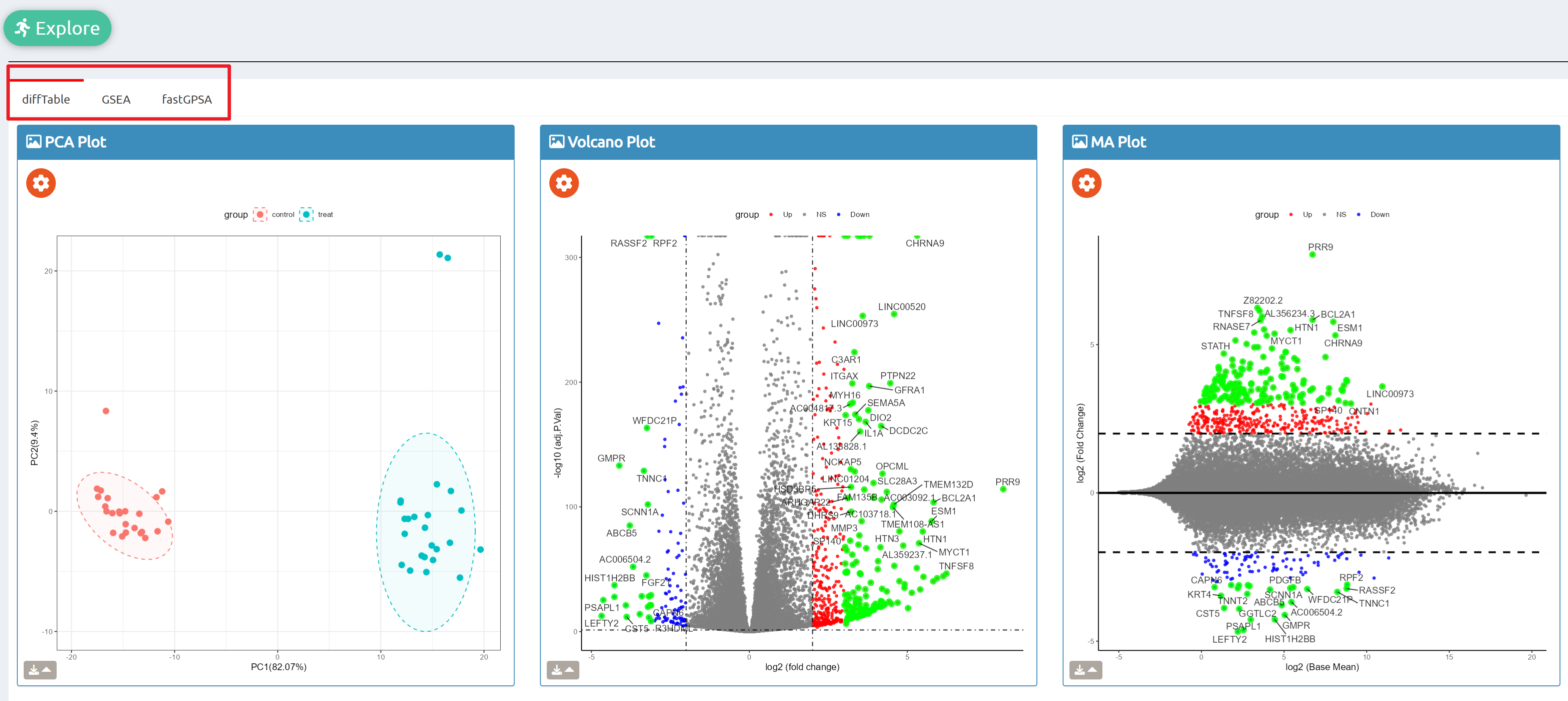
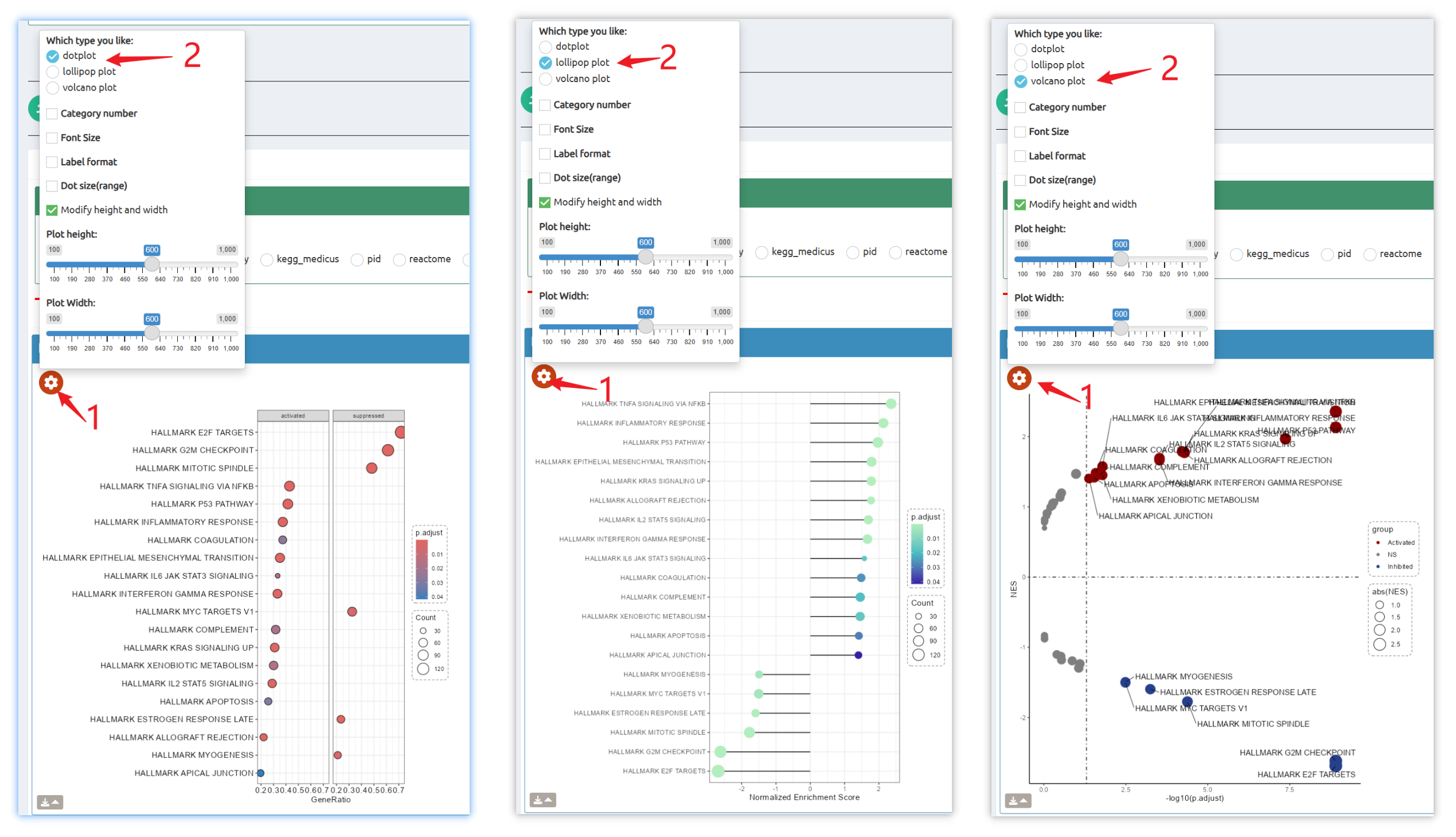
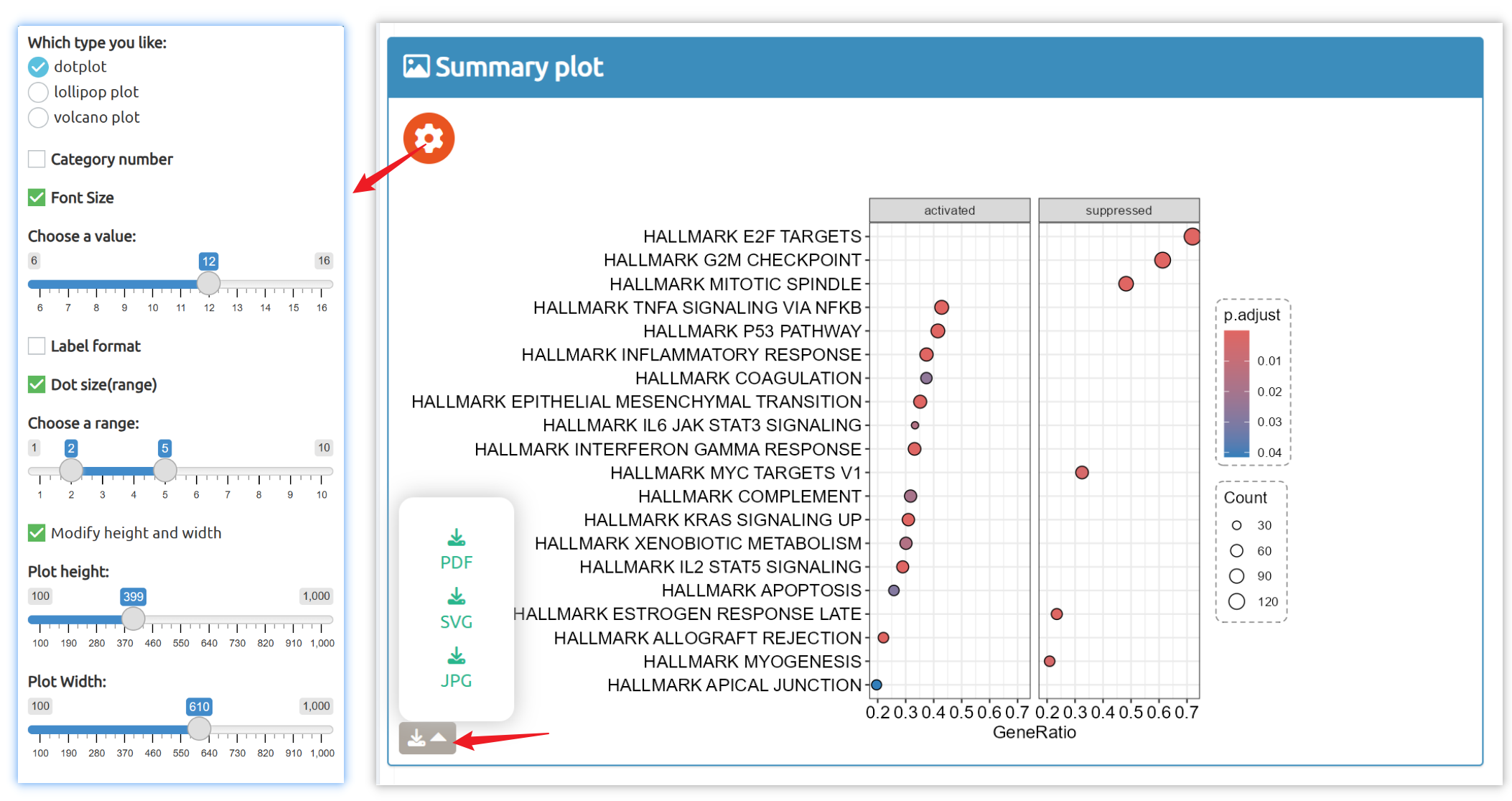
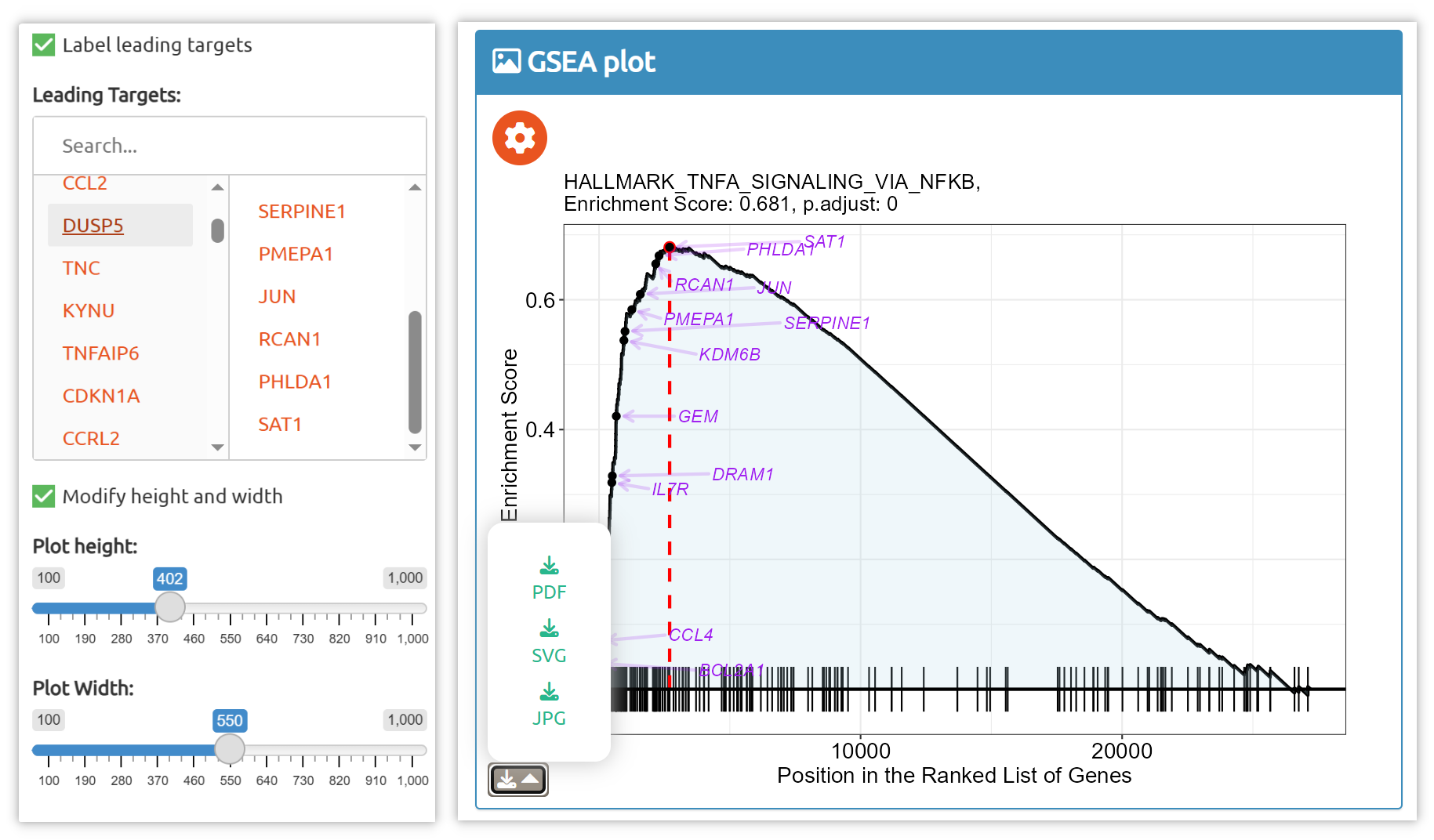
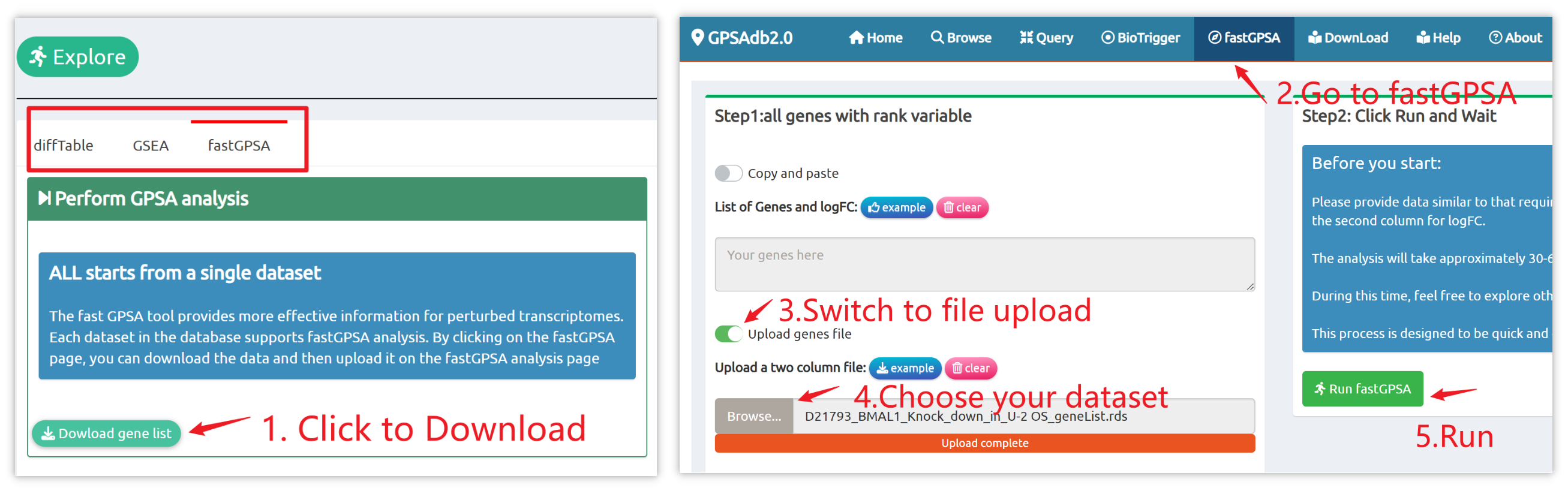
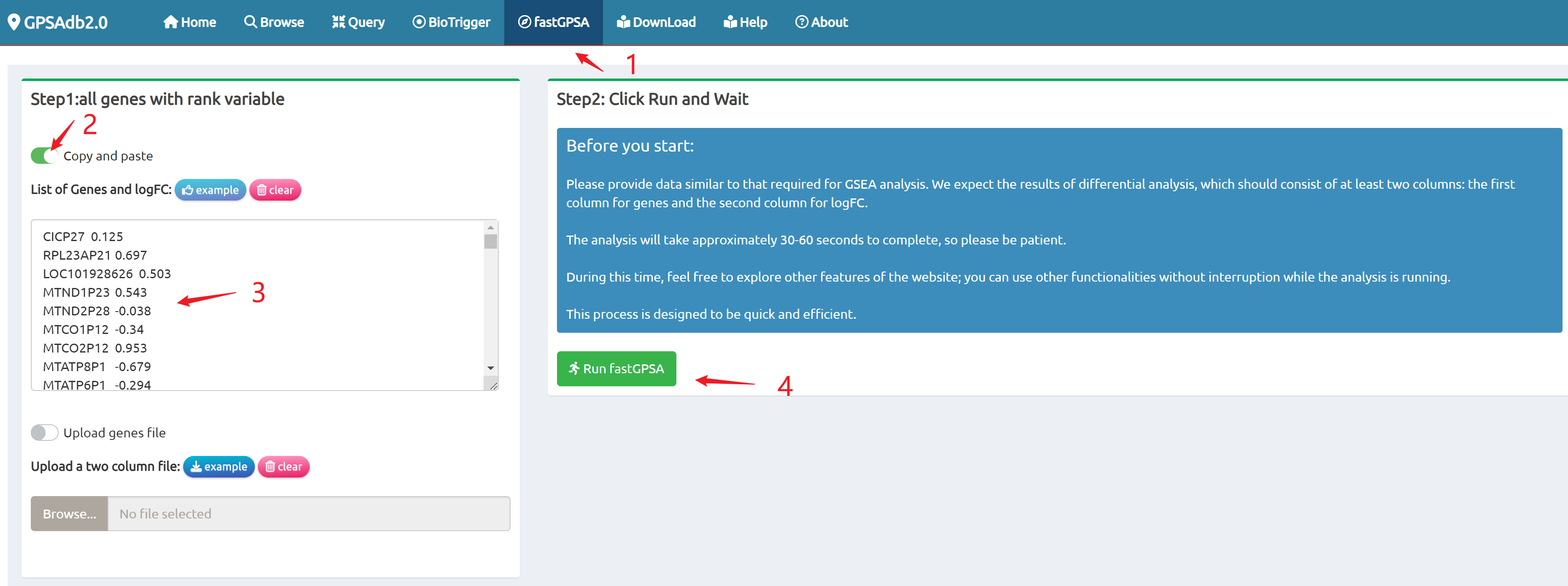
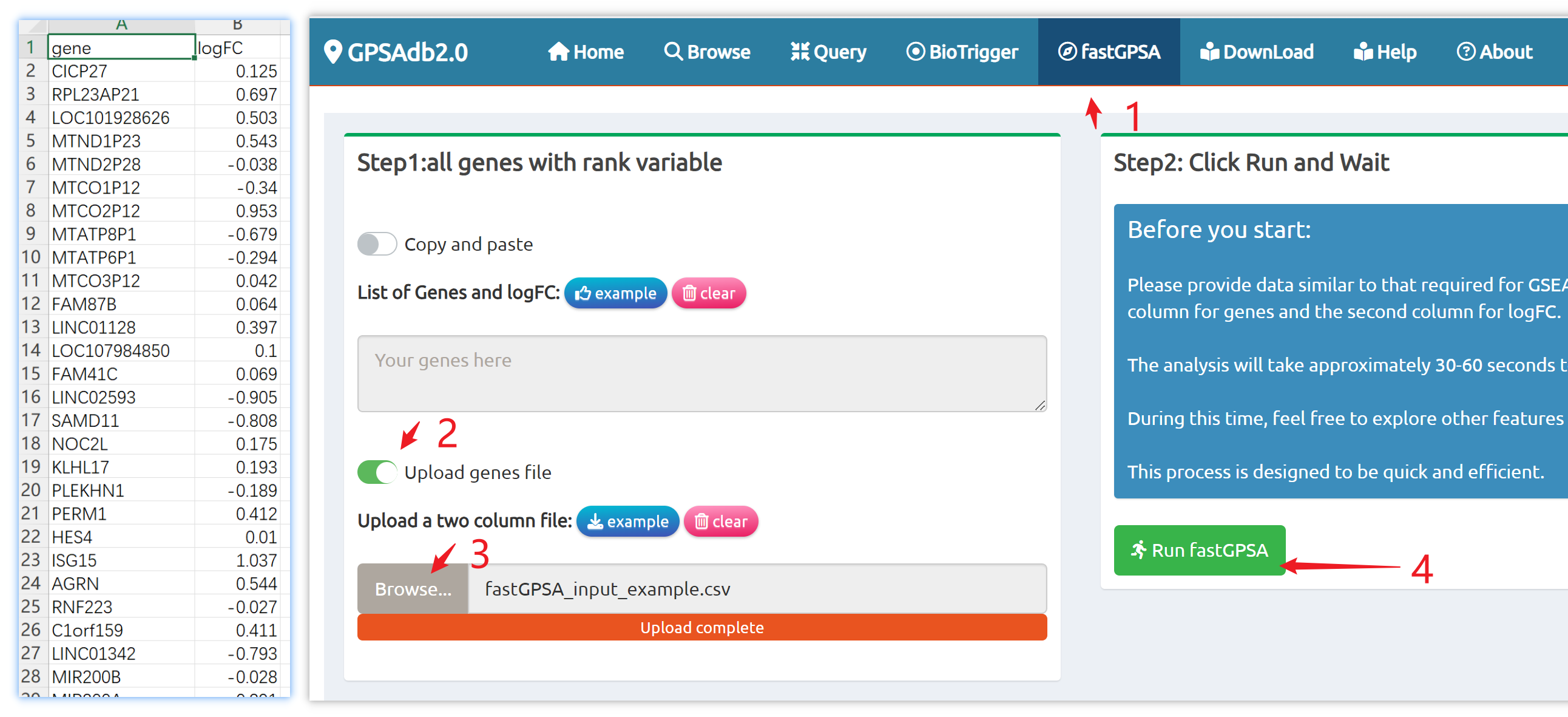
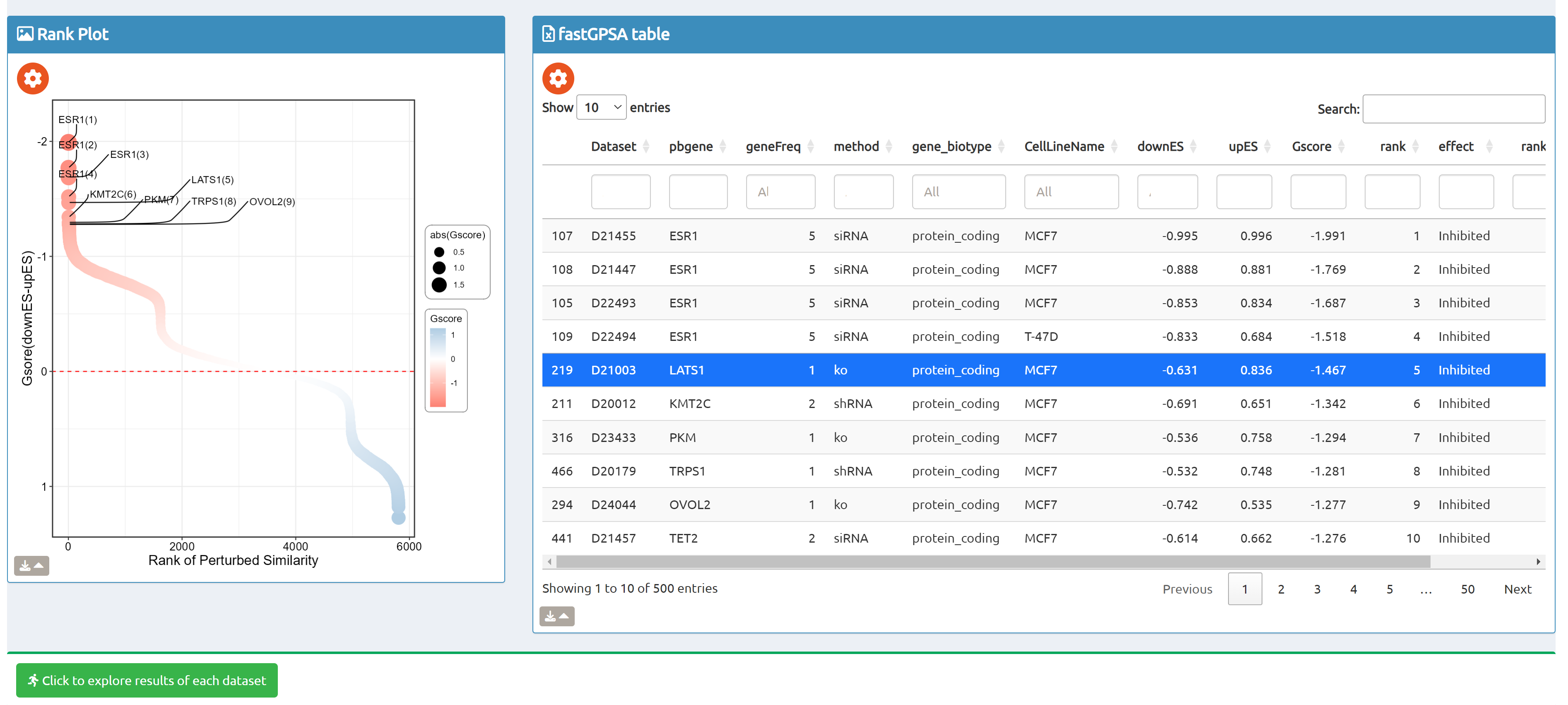
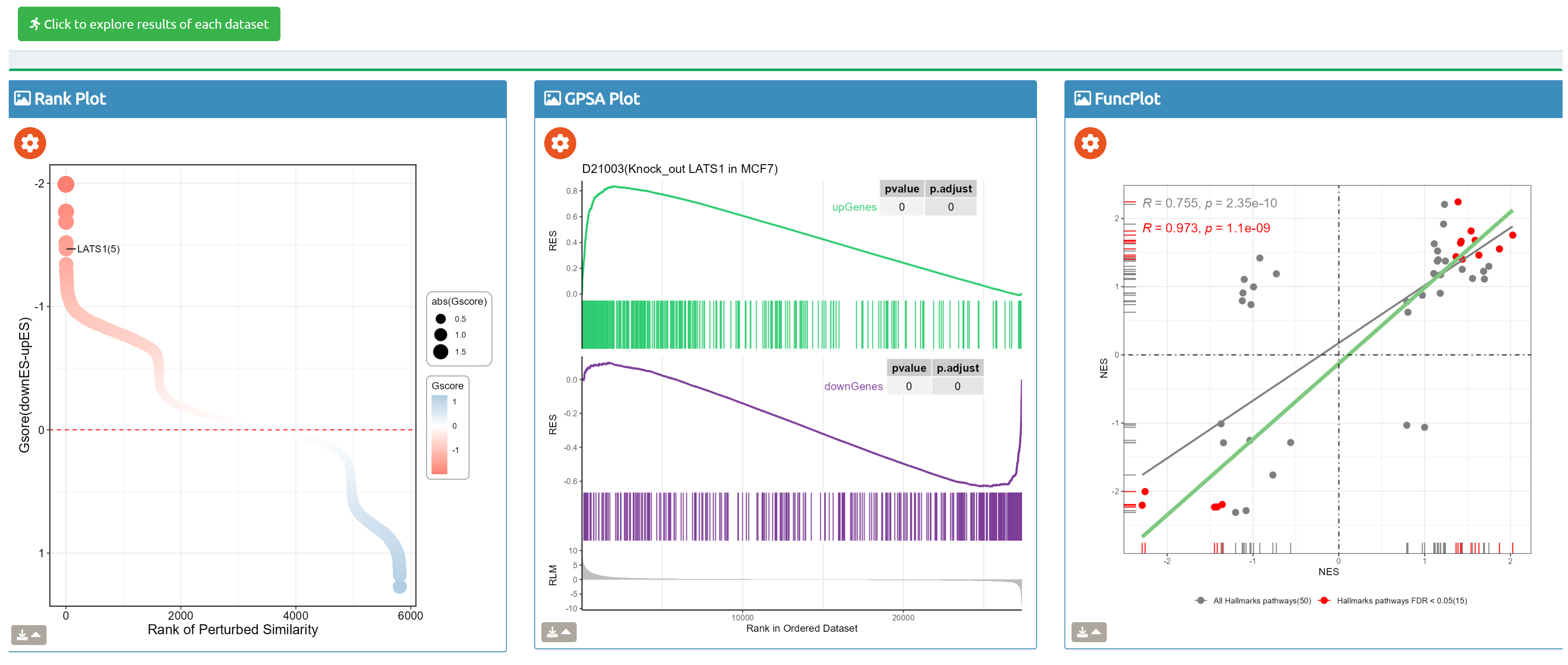
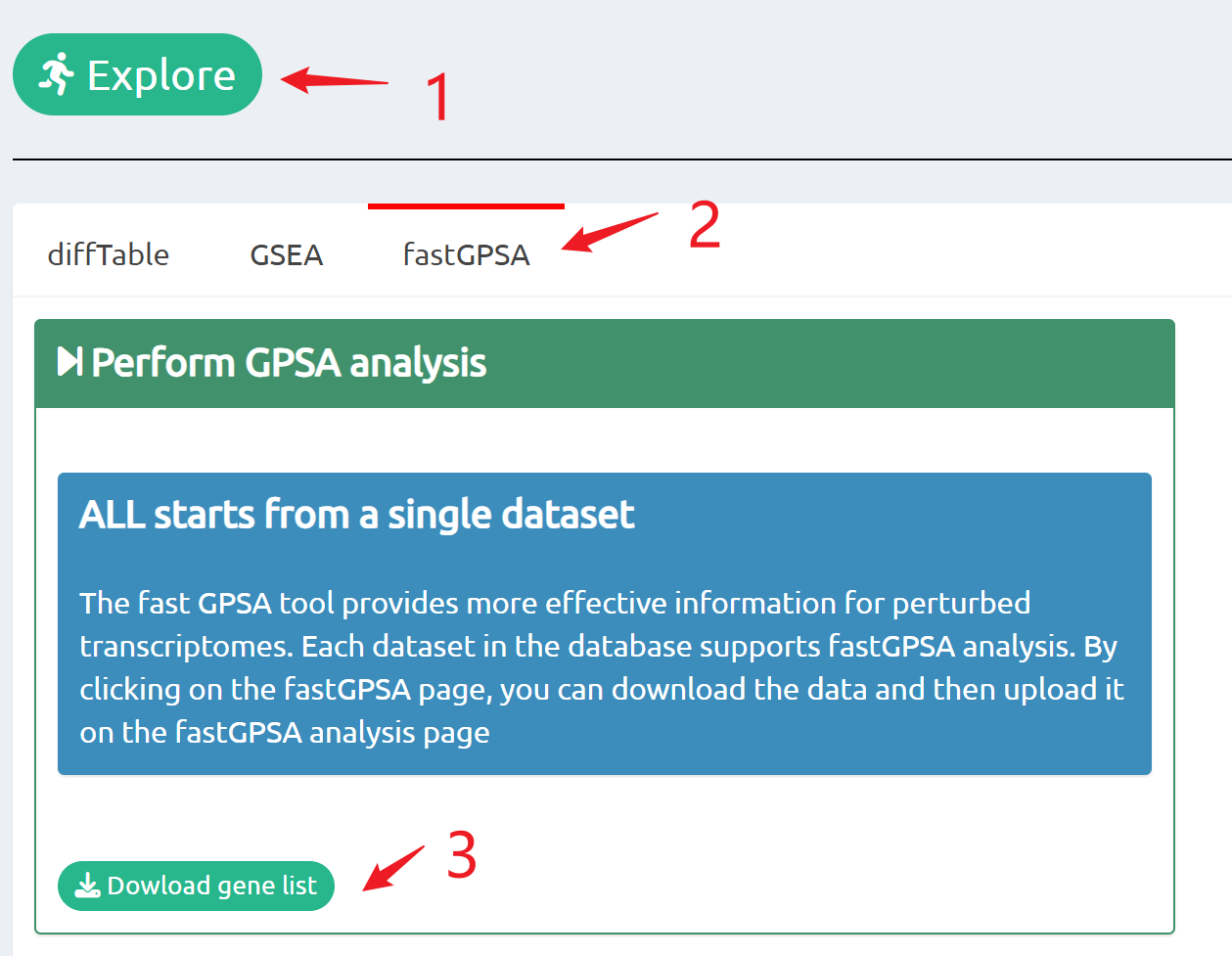
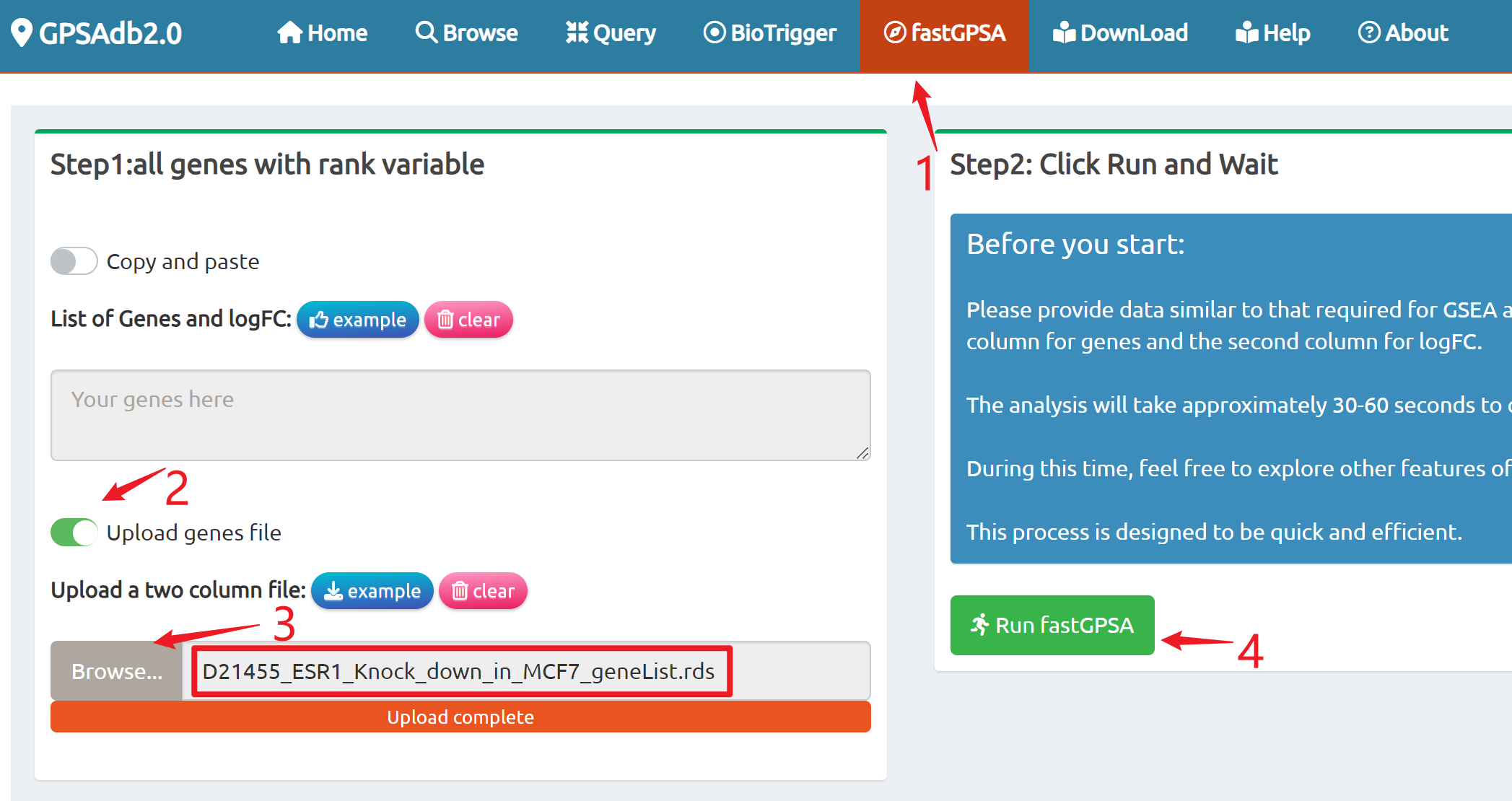
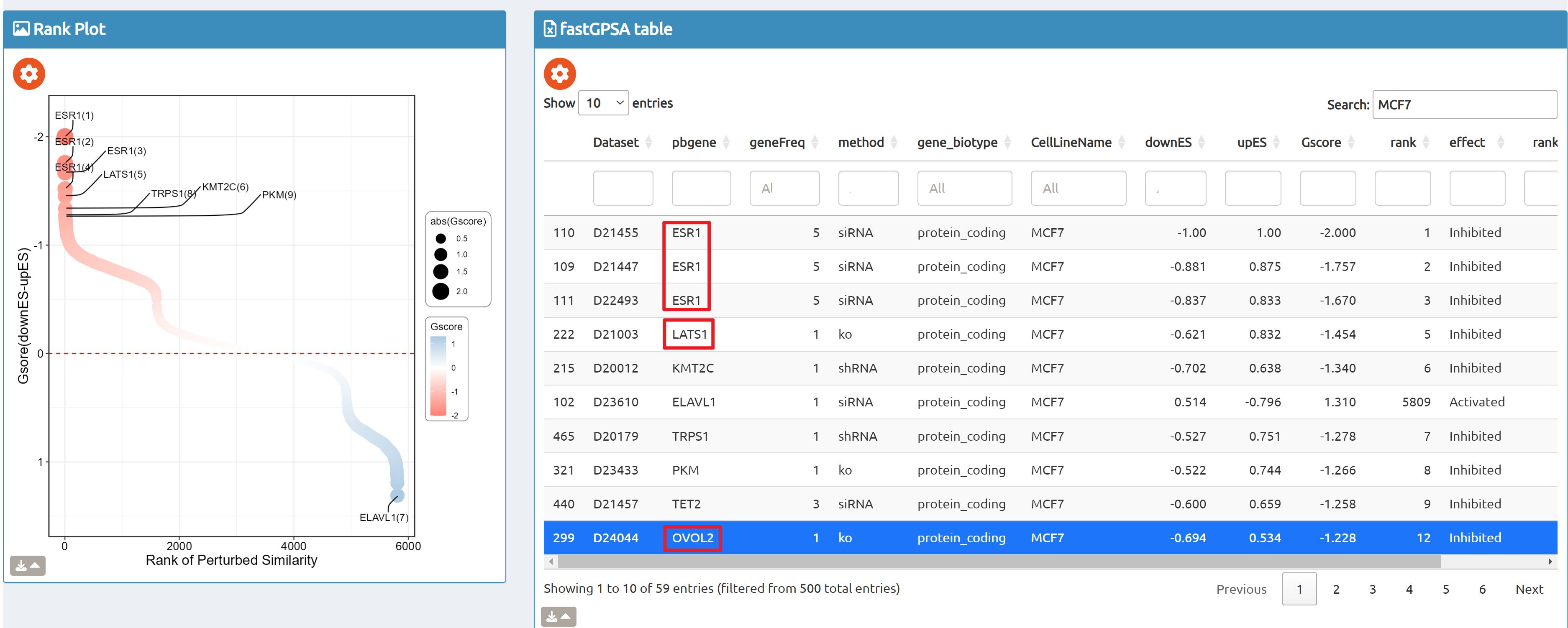
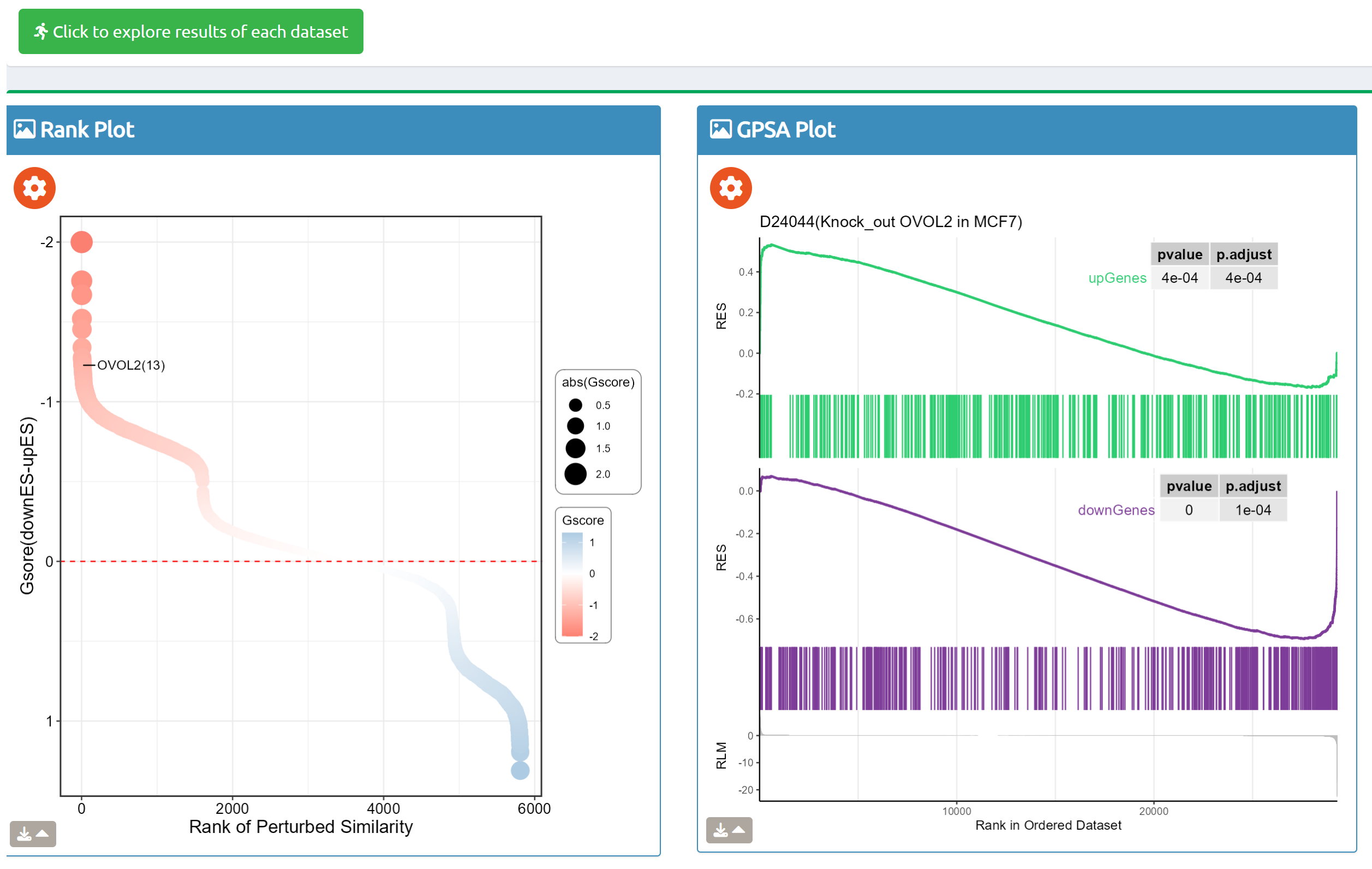
- fastGPSA: Upload a differential-expression table; fastGPSA performs bidirectional enrichment (up/down) and ranks matched perturbations via a G-score, with interactive visuals (Rank Plot, GPSAPlot).
- Reproducible results: One-click access to datasets, consolidated downloads, and step-by-step case studies to guide typical research workflows.
Use cases: Discover regulatory genes, illuminate disease mechanisms, and prioritize potential therapeutic targets from perturbation transcriptomes—quickly, transparently, and at scale.
Browse
7,665 datasets to explore
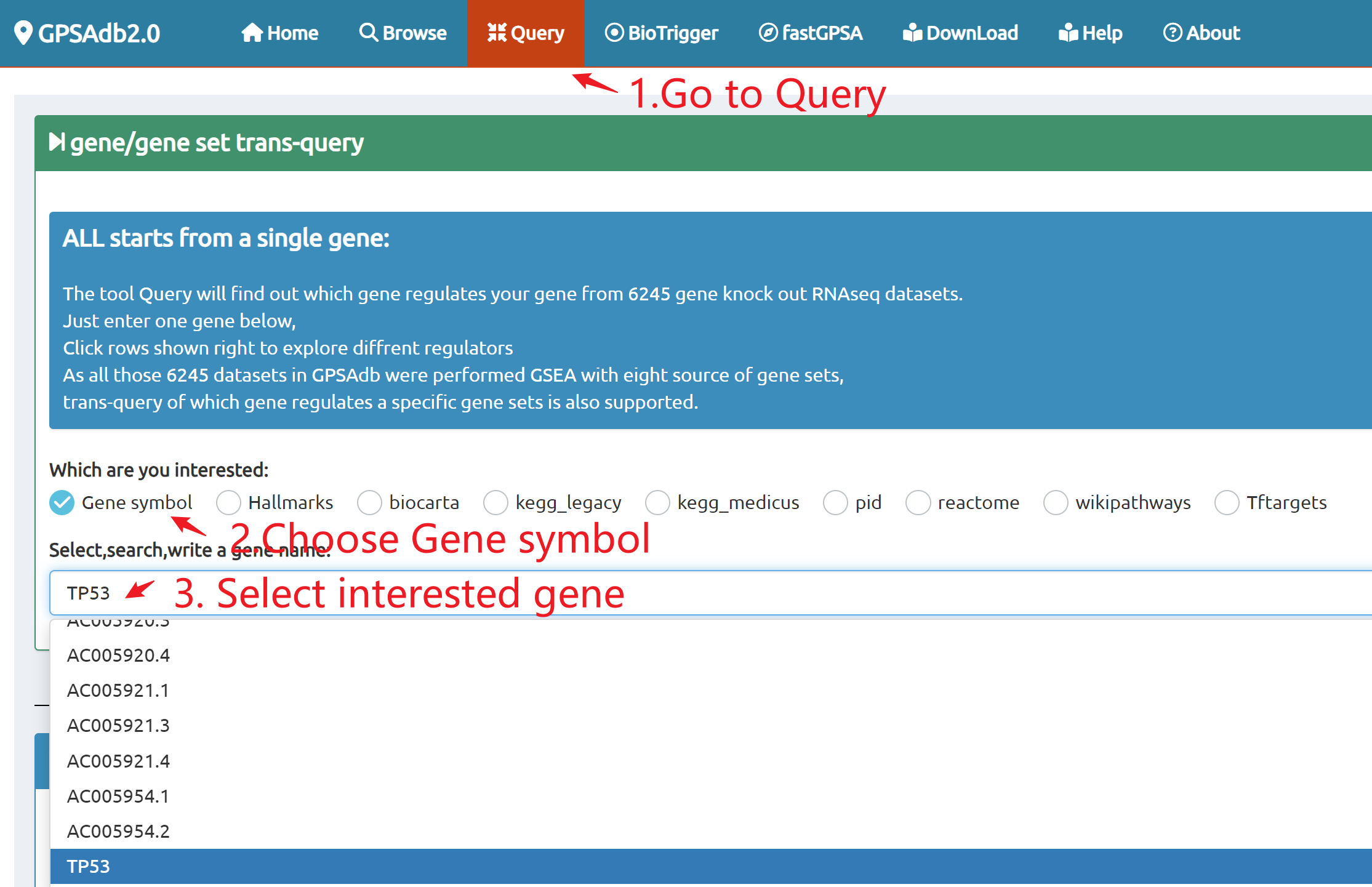
Query
Query by single gene/gene set
BioTrigger
Find triggers of all gene sets
fastGPSA
Enrich analysis with your interested genes
Download
Download all data easily
About
More information
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.